Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population
- PMID: 31199554
- PMCID: PMC6920336
- DOI: 10.1111/pbi.13191
Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population
Abstract
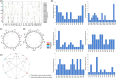
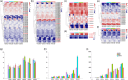
Cotton is widely cultivated globally because it provides natural fibre for the textile industry and human use. To identify quantitative trait loci (QTLs)/genes associated with fibre quality and yield, a recombinant inbred line (RIL) population was developed in upland cotton. A consensus map covering the whole genome was constructed with three types of markers (8295 markers, 5197.17 centimorgans (cM)). Six fibre yield and quality traits were evaluated in 17 environments, and 983 QTLs were identified, 198 of which were stable and mainly distributed on chromosomes 4, 6, 7, 13, 21 and 25. Thirty-seven QTL clusters were identified, in which 92.8% of paired traits with significant medium or high positive correlations had the same QTL additive effect directions, and all of the paired traits with significant medium or high negative correlations had opposite additive effect directions. In total, 1297 genes were discovered in the QTL clusters, 414 of which were expressed in two RNA-Seq data sets. Many genes were discovered, 23 of which were promising candidates. Six important QTL clusters that included both fibre quality and yield traits were identified with opposite additive effect directions, and those on chromosome 13 (qClu-chr13-2) could increase fibre quality but reduce yield; this result was validated in a natural population using three markers. These data could provide information about the genetic basis of cotton fibre quality and yield and help cotton breeders to improve fibre quality and yield simultaneously.
Keywords: QTL clusters; consensus genetic map; fibre quality; fibre yield; gene expression level; genetic correlation; upland cotton.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Cai, H.W. and Morishima, H. (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104, 1217–1228. - PubMed
-
- Cai, C.P. , Niu, E.L. , Du, H. , Zhao, L. , Feng, Y. and Guo, W.Z. (2014) Genome‐wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J. 2, 87–101.
-
- Cao, Z.B. , Zhu, X.F. , Chen, H. and Zhang, T.Z. (2015) Fine mapping of clustered quantitative trait loci for fiber quality on chromosome 7 using a Gossypium barbadense introgressed line. Mol. Breed. 35, 1–13.
-
- Chandnani, R. , Kim, C. , Guo, H. , Shehzad, T. , Wallace, J.G. , He, D.H. , Zhang, Z.S. et al. (2018) Genetic analysis of gossypium fiber quality traits in reciprocal advanced backcross populations. Plant Genome. 11, 170057. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

