Identification of potential diagnostic biomarkers for Parkinson's disease
- PMID: 31199560
- PMCID: PMC6668373
- DOI: 10.1002/2211-5463.12687
Identification of potential diagnostic biomarkers for Parkinson's disease
Abstract
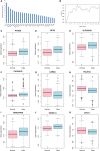
The identification of biomarkers for early diagnosis of Parkinson's disease (PD) prior to the onset of symptoms may improve the effectiveness of therapy. To identify potential biomarkers, we downloaded microarray datasets of PD from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) between PD and normal control (NC) groups were obtained, and the feature selection procedure and classification model were used to identify optimal diagnostic gene biomarkers for PD. A total of 1229 genes (640 up-regulated and 589 down-regulated) were obtained for PD, and nine DEGs (PTGDS, GPX3, SLC25A20, CACNA1D, LRRN3, POLR1D, ARHGAP26, TNFSF14 and VPS11) were selected as optimal PD biomarkers with great diagnostic value. These nine DEGs were significantly enriched in regulation of circadian sleep/wake cycle, sleep and gonadotropin-releasing hormone signaling pathway. Finally, we examined the expression of GPX3, SLC25A20, LRRN3 and POLR1D in blood samples of patients with PD by qRT-PCR. GPX3, LRRN3 and POLR1D exhibited the same expression pattern as in our analysis. In conclusion, this study identified nine DEGs that may serve as potential biomarkers of PD.
Keywords: Parkinson's disease; biomarker; differentially expressed genes; integrated analysis.
© 2019 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

