Characterisation of HvVIP1 and expression profile analysis of stress response regulators in barley under Agrobacterium and Fusarium infections
- PMID: 31199821
- PMCID: PMC6570034
- DOI: 10.1371/journal.pone.0218120
Characterisation of HvVIP1 and expression profile analysis of stress response regulators in barley under Agrobacterium and Fusarium infections
Abstract
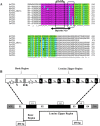
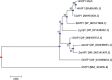
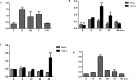
Arabidopsis thaliana's VirE2-Interacting Protein 1 (VIP1) interacts with Agrobacterium tumefaciens VirE2 protein and regulates stress responses and plant immunity signaling occurring downstream of the Mitogen-Activated Protein Kinase (MPK3) signal transduction pathway. In this study, a full-length cDNA of 972bp encoding HvVIP1 was obtained from barley (Hordeum vulgare L.) leaves. A corresponding 323 amino acid poly-peptide was shown to carry the conserved bZIP (Basic Leucine Zipper) domain within its 157th and 223rd amino acid residue. 13 non-synonymous SNPs were spotted within the HvVIP1 bZIP domain sequence when compared with AtVIP1. Moreover, minor differences in the bZIP domain locations and lengths were noted when comparing Arabidopsis thaliana and Hordeum vulgare VIP1 proteins through the 3D models, structural domain predictions and disorder prediction profiling. The expression of HvVIP1 was stable in barley tissues infected by pathogen (whether Agrobacterium tumefaciens or Fusarium culmorum), but was induced at specific time points. We found a strong correlation between the transcript accumulation of HvVIP1 and barley PR- genes HvPR1, HvPR4 and HvPR10, but not with HvPR3 and HvPR5, probably due to low induction of those particular genes. In addition, a gene encoding for a member of the barley MAPK family, HvMPK1, showed significantly higher expression after pathogenic infection of barley cells. Collectively, our results might suggest that early expression of PR genes upon infection in barley cells play a pivotal role in the Agrobacterium-resistance of this plant.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Transcriptome analysis of the barley-Fusarium graminearum interaction.Mol Plant Microbe Interact. 2006 Apr;19(4):407-17. doi: 10.1094/MPMI-19-0407. Mol Plant Microbe Interact. 2006. PMID: 16610744
-
Proteome analysis of Fusarium head blight in grains of naked barley (Hordeum vulgare subsp. nudum).Proteomics. 2011 Mar;11(5):972-85. doi: 10.1002/pmic.201000322. Epub 2011 Jan 27. Proteomics. 2011. PMID: 21271677
-
The Agrobacterium VirE2 effector interacts with multiple members of the Arabidopsis VIP1 protein family.Mol Plant Pathol. 2018 May;19(5):1172-1183. doi: 10.1111/mpp.12595. Epub 2017 Oct 24. Mol Plant Pathol. 2018. PMID: 28802023 Free PMC article.
-
Brassinosteroid enhances resistance to fusarium diseases of barley.Phytopathology. 2013 Dec;103(12):1260-7. doi: 10.1094/PHYTO-05-13-0111-R. Phytopathology. 2013. PMID: 23777406
-
VIP1: linking Agrobacterium-mediated transformation to plant immunity?Plant Cell Rep. 2010 Aug;29(8):805-12. doi: 10.1007/s00299-010-0870-4. Epub 2010 May 15. Plant Cell Rep. 2010. PMID: 20473505 Review.
Cited by
-
TALEN-Based HvMPK3 Knock-Out Attenuates Proteome and Root Hair Phenotypic Responses to flg22 in Barley.Front Plant Sci. 2021 Apr 29;12:666229. doi: 10.3389/fpls.2021.666229. eCollection 2021. Front Plant Sci. 2021. PMID: 33995462 Free PMC article.
-
Agrobacterium-mediated gene transfer: recent advancements and layered immunity in plants.Planta. 2022 Jul 11;256(2):37. doi: 10.1007/s00425-022-03951-x. Planta. 2022. PMID: 35819629 Free PMC article. Review.
-
Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear (Pyrus breschneideri).BMC Plant Biol. 2021 Dec 9;21(1):583. doi: 10.1186/s12870-021-03356-0. BMC Plant Biol. 2021. PMID: 34886805 Free PMC article.
-
Pseudocercospora fijiensis Conidial Germination Is Dominated by Pathogenicity Factors and Effectors.J Fungi (Basel). 2023 Sep 27;9(10):970. doi: 10.3390/jof9100970. J Fungi (Basel). 2023. PMID: 37888226 Free PMC article.
References
-
- Schell J, Van Montagu M, De Beuckeleer M, De Block M, Depicker A, De Wilde M, et al. Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1979; 204, 1155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

