Defining genotype-phenotype relationships in patients with hypertrophic cardiomyopathy using cardiovascular magnetic resonance imaging
- PMID: 31199839
- PMCID: PMC6568393
- DOI: 10.1371/journal.pone.0217612
Defining genotype-phenotype relationships in patients with hypertrophic cardiomyopathy using cardiovascular magnetic resonance imaging
Abstract
Purpose: HCM is the most common inherited cardiomyopathy. Historically, there has been poor correlation between genotype and phenotype. However, CMR has the potential to more accurately assess disease phenotype. We characterized phenotype with CMR in a cohort of patients with confirmed HCM and high prevalence of genetic testing.
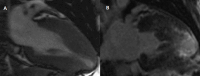
Methods: Patients with a diagnosis of HCM, who had undergone contrast-enhanced CMR were identified. Left ventricular mass index (LVMI) and volumes were measured from steady-state free precession sequences. Late gadolinium enhancement (LGE) was quantified using the full width, half maximum method. All patients were prospectively followed for the development of septal reduction therapy, arrhythmia or death.
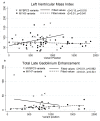
Results: We included 273 patients, mean age 51.2 ± 15.5, 62.9% male. Of those patients 202 (74.0%) underwent genetic testing with 90 pathogenic, likely pathogenic, or rare variants and 13 variants of uncertain significance identified. Median follow-up was 1138 days. Mean LVMI was 82.7 ± 30.6 and 145 patients had late gadolinium enhancement (LGE). Patients with beta-myosin heavy chain (MYH7) mutations had higher LV ejection fraction (68.8 vs 59.1, p<0.001) than those with cardiac myosin binding protein C (MYBPC3) mutations. Patients with MYBPC3 mutations were more likely to have LVEF < 55% (29.7% vs 4.9%, p = 0.005) or receive a defibrillator than those with MYH7 mutations (54.1% vs 26.8%, p = 0.020).
Conclusions: We found that patients with MYBPC3 mutations were more likely to have impaired ventricular function and may be more prone to arrhythmic events. Larger studies using CMR phenotyping may be capable of identifying additional characteristics associated with less frequent genetic causes of HCM.
Conflict of interest statement
The authors have read the journal's policy and the authors of this manuscript have the following competing interests: RJHM is supported by the Arthur J E Child fellowship grant. GE Healthcare provides equipment research support to PCY, SH. AP was supported by the Breetwor Foundation. AS was supported by a research grant to EAA from MyoKardia. EAA is a consultant and receives researching funding with Myokardia. MTW has ownership interest and is a stockholder in Personalis and has ownership interest, research funding and consulting with MyoKardia. There are no patents, products in development or marketed products associated with this research to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures

References
-
- Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–9. - PubMed
-
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001. February 23;104(4):557–67. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

