Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery
- PMID: 31199850
- PMCID: PMC6594655
- DOI: 10.1371/journal.ppat.1007842
Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery
Abstract
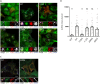
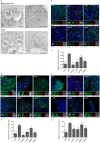
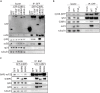
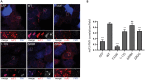
G3BP-1 and -2 (hereafter referred to as G3BP) are multifunctional RNA-binding proteins involved in stress granule (SG) assembly. Viruses from diverse families target G3BP for recruitment to replication or transcription complexes in order to block SG assembly but also to acquire pro-viral effects via other unknown functions of G3BP. The Old World alphaviruses, including Semliki Forest virus (SFV) and chikungunya virus (CHIKV) recruit G3BP into viral replication complexes, via an interaction between FGDF motifs in the C-terminus of the viral non-structural protein 3 (nsP3) and the NTF2-like domain of G3BP. To study potential proviral roles of G3BP, we used human osteosarcoma (U2OS) cell lines lacking endogenous G3BP generated using CRISPR-Cas9 and reconstituted with a panel of G3BP1 mutants and truncation variants. While SFV replicated with varying efficiency in all cell lines, CHIKV could only replicate in cells expressing G3BP1 variants containing both the NTF2-like and the RGG domains. The ability of SFV to replicate in the absence of G3BP allowed us to study effects of different domains of the protein. We used immunoprecipitation to demonstrate that that both NTF2-like and RGG domains are necessary for the formation a complex between nsP3, G3BP1 and the 40S ribosomal subunit. Electron microscopy of SFV-infected cells revealed that formation of nsP3:G3BP1 complexes via the NTF2-like domain was necessary for clustering of cytopathic vacuoles (CPVs) and that the presence of the RGG domain was necessary for accumulation of electron dense material containing G3BP1 and nsP3 surrounding the CPV clusters. Clustered CPVs also exhibited localised high levels of translation of viral mRNAs as detected by ribopuromycylation staining. These data confirm that G3BP is a ribosomal binding protein and reveal that alphaviral nsP3 uses G3BP to concentrate viral replication complexes and to recruit the translation initiation machinery, promoting the efficient translation of viral mRNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

