Rab32 GTPase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation
- PMID: 31199852
- PMCID: PMC6594657
- DOI: 10.1371/journal.ppat.1007879
Rab32 GTPase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation
Abstract
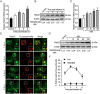
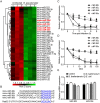
Burkholderia pseudomallei is a gram-negative, facultative intracellular bacterium, which causes a disease known as melioidosis. Professional phagocytes represent a crucial first line of innate defense against invading pathogens. Uptake of pathogens by these cells involves the formation of a phagosome that matures by fusing with early and late endocytic vesicles, resulting in killing of ingested microbes. Host Rab GTPases are central regulators of vesicular trafficking following pathogen phagocytosis. However, it is unclear how Rab GTPases interact with B. pseudomallei to regulate the transport and maturation of bacterial-containing phagosomes. Here, we showed that the host Rab32 plays an important role in mediating antimicrobial activity by promoting phagosome maturation at an early phase of infection with B. pseudomallei. And we demonstrated that the expression level of Rab32 is increased through the downregulation of the synthesis of miR-30b/30c in B. pseudomallei infected macrophages. Subsequently, we showed that B. pseudomallei resides temporarily in Rab32-positive compartments with late endocytic features. And Rab32 enhances phagosome acidification and promotes the fusion of B. pseudomallei-containing phagosomes with lysosomes to activate cathepsin D, resulting in restricted intracellular growth of B. pseudomallei. Additionally, Rab32 mediates phagosome maturation depending on its guanosine triphosphate/guanosine diphosphate (GTP/GDP) binding state. Finally, we report the previously unrecognized role of miR-30b/30c in regulating B. pseudomallei-containing phagosome maturation by targeting Rab32 in macrophages. Altogether, we provide a novel insight into the host immune-regulated cellular pathway against B. pseudomallei infection is partially dependent on Rab32 trafficking pathway, which regulates phagosome maturation and enhances the killing of this bacterium in macrophages.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Burkholderia pseudomallei BopE suppresses the Rab32-dependent defense pathway to promote its intracellular replication and virulence.mSphere. 2024 Nov 21;9(11):e0045324. doi: 10.1128/msphere.00453-24. Epub 2024 Oct 21. mSphere. 2024. PMID: 39431830 Free PMC article.
-
Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes.Traffic. 2011 Apr;12(4):407-20. doi: 10.1111/j.1600-0854.2011.01165.x. Epub 2011 Feb 21. Traffic. 2011. PMID: 21255211
-
An ultrastructural study of the phagocytosis of Burkholderia pseudomallei.Microbios. 1998;94(377):35-45. Microbios. 1998. PMID: 9785484
-
Exploitation of host cells by Burkholderia pseudomallei.Int J Med Microbiol. 2004 Apr;293(7-8):549-55. doi: 10.1078/1438-4221-00292. Int J Med Microbiol. 2004. PMID: 15149030 Review.
-
Autophagy and burkholderia.Immunol Cell Biol. 2015 Jan;93(1):18-24. doi: 10.1038/icb.2014.87. Epub 2014 Oct 21. Immunol Cell Biol. 2015. PMID: 25331551 Review.
Cited by
-
VARP and Rab9 Are Dispensable for the Rab32/BLOC-3 Dependent Salmonella Killing.Front Cell Infect Microbiol. 2020 Dec 16;10:581024. doi: 10.3389/fcimb.2020.581024. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33392103 Free PMC article.
-
The Rab32/BLOC-3-dependent pathway mediates host defense against different pathogens in human macrophages.Sci Adv. 2021 Jan 15;7(3):eabb1795. doi: 10.1126/sciadv.abb1795. Print 2021 Jan. Sci Adv. 2021. PMID: 33523895 Free PMC article.
-
Integrative p53, micro-RNA and Cathepsin Protease Co-Regulatory Expression Networks in Cancer.Cancers (Basel). 2020 Nov 20;12(11):3454. doi: 10.3390/cancers12113454. Cancers (Basel). 2020. PMID: 33233599 Free PMC article. Review.
-
Regulatory Network Analysis in Estradiol-Treated Human Endothelial Cells.Int J Mol Sci. 2021 Jul 30;22(15):8193. doi: 10.3390/ijms22158193. Int J Mol Sci. 2021. PMID: 34360960 Free PMC article.
-
MiR-124a Regulates Extracellular Vesicle Release by Targeting GTPase Rabs in Lung Cancer.Front Oncol. 2020 Aug 20;10:1454. doi: 10.3389/fonc.2020.01454. eCollection 2020. Front Oncol. 2020. PMID: 32974168 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

