Study of conformational transitions of i-motif DNA using time-resolved fluorescence and multivariate analysis methods
- PMID: 31199873
- PMCID: PMC6649798
- DOI: 10.1093/nar/gkz522
Study of conformational transitions of i-motif DNA using time-resolved fluorescence and multivariate analysis methods
Abstract
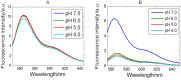
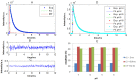
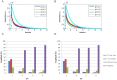
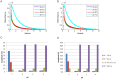
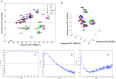
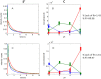
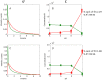
Recently, the presence of i-motif structures at C-rich sequences in human cells and their regulatory functions have been demonstrated. Despite numerous steady-state studies on i-motif at neutral and slightly acidic pH, the number and nature of conformation of this biological structure are still controversial. In this work, the fluorescence lifetime of labelled molecular beacon i-motif-forming DNA sequences at different pH values is studied. The influence of the nature of bases at the lateral loops and the presence of a Watson-Crick-stabilized hairpin are studied by means of time-correlated single-photon counting technique. This allows characterizing the existence of several conformers for which the fluorophore has lifetimes ranging from picosecond to nanosecond. The information on the existence of different i-motif structures at different pH values has been obtained by the combination of classical global decay fitting of fluorescence traces, which provides lifetimes associated with the events defined by the decay of each sequence and multivariate analysis, such as principal component analysis or multivariate curve resolution based on alternating least squares. Multivariate analysis, which is seldom used for this kind of data, was crucial to explore similarities and differences of behaviour amongst the different DNA sequences and to model the presence and identity of the conformations involved in the pH range of interest. The results point that, for i-motif, the intrachain contact formation and its dissociation show lifetimes ten times faster than for the open form of DNA sequences. They also highlight that the presence of more than one i-motif species for certain DNA sequences according to the length of the sequence and the composition of the bases in the lateral loop.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Bacolla A., Wells R.D.. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009; 48:273–285. - PubMed
-
- Bucek P., Jaumot J., Aviñó A., Eritja R., Gargallo R.. PH-modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chem. A Eur. J. 2009; 15:12663–12671. - PubMed
-
- Benabou S., Ferreira R., Aviñó A., González C., Lyonnais S., Solà M., Eritja R., Jaumot J., Gargallo R.. Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops. Biochim. Biophys. Acta Gen. Subj. 2014; 1840:41–52. - PubMed

