Autophagy-Independent Functions of the Autophagy Machinery
- PMID: 31199916
- PMCID: PMC7173070
- DOI: 10.1016/j.cell.2019.05.026
Autophagy-Independent Functions of the Autophagy Machinery
Abstract
Macroautophagy (herein referred to as autophagy) is an evolutionary ancient mechanism that culminates with the lysosomal degradation of superfluous or potentially dangerous cytosolic entities. Over the past 2 decades, the molecular mechanisms underlying several variants of autophagy have been characterized in detail. Accumulating evidence suggests that most, if not all, components of the molecular machinery for autophagy also mediate autophagy-independent functions. Here, we discuss emerging data on the non-autophagic functions of autophagy-relevant proteins.
Keywords: ATG5; BECN1; LC3-associated phagocytosis; proliferation; regulated cell death; vesicular trafficking.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
L.G. provides remunerated consulting to OmniSEQ (Buffalo, NY, USA), Astra Zeneca (Gaithersburg, MD, USA), VL47 (New York, NY, USA), and the Luke Heller TECPR2 Foundation (Boston, MA, USA), and he is member of the Scientific Advisory Committee of OmniSEQ (Buffalo, NY, USA). D.R.G. has no conflicts of interest to declare.
Figures
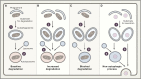

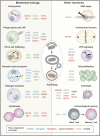
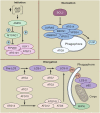
Similar articles
-
Cargo recognition and degradation by selective autophagy.Nat Cell Biol. 2018 Mar;20(3):233-242. doi: 10.1038/s41556-018-0037-z. Epub 2018 Feb 23. Nat Cell Biol. 2018. PMID: 29476151 Free PMC article. Review.
-
Lipids and Lipid-Binding Proteins in Selective Autophagy.J Mol Biol. 2020 Jan 3;432(1):135-159. doi: 10.1016/j.jmb.2019.05.051. Epub 2019 Jun 14. J Mol Biol. 2020. PMID: 31202884 Review.
-
Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9.Cell Rep. 2017 Sep 5;20(10):2341-2356. doi: 10.1016/j.celrep.2017.08.034. Cell Rep. 2017. PMID: 28877469 Free PMC article.
-
A current view of molecular dissection in autophagy machinery.J Physiol Biochem. 2020 Aug;76(3):357-372. doi: 10.1007/s13105-020-00746-0. Epub 2020 May 25. J Physiol Biochem. 2020. PMID: 32451934 Review.
-
Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration.Autophagy. 2019 Apr;15(4):631-651. doi: 10.1080/15548627.2018.1535292. Epub 2018 Nov 5. Autophagy. 2019. PMID: 30335591 Free PMC article.
Cited by
-
ALK inhibition activates LC3B-independent, protective autophagy in EML4-ALK positive lung cancer cells.Sci Rep. 2021 Apr 27;11(1):9011. doi: 10.1038/s41598-021-87966-6. Sci Rep. 2021. PMID: 33907223 Free PMC article.
-
Complement C3 deposition restricts the proliferation of internalized Staphylococcus aureus by promoting autophagy.Front Cell Infect Microbiol. 2024 Sep 6;14:1400068. doi: 10.3389/fcimb.2024.1400068. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39310788 Free PMC article.
-
Renal tubular epithelial cell quality control mechanisms as therapeutic targets in renal fibrosis.J Pharm Anal. 2024 Aug;14(8):100933. doi: 10.1016/j.jpha.2024.01.001. Epub 2024 Jan 3. J Pharm Anal. 2024. PMID: 39247486 Free PMC article. Review.
-
LC3-associated endocytosis and the functions of Rubicon and ATG16L1.Sci Adv. 2022 Oct 28;8(43):eabo5600. doi: 10.1126/sciadv.abo5600. Epub 2022 Oct 26. Sci Adv. 2022. PMID: 36288306 Free PMC article. Review.
-
Reductive damage induced autophagy inhibition for tumor therapy.Nano Res. 2023;16(4):5226-5236. doi: 10.1007/s12274-022-5139-z. Epub 2022 Nov 22. Nano Res. 2023. PMID: 36465522 Free PMC article.
References
-
- Al-Younes H.M., Al-Zeer M.A., Khalil H., Gussmann J., Karlas A., Machuy N., Brinkmann V., Braun P.R., Meyer T.F. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy. 2011;7:814–828. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

