Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus
- PMID: 31200111
- PMCID: PMC6876278
- DOI: 10.1016/j.meegid.2019.103917
Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus
Abstract
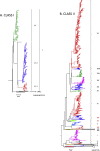
Several Avian paramyxoviruses 1 (synonymous with Newcastle disease virus or NDV, used hereafter) classification systems have been proposed for strain identification and differentiation. These systems pioneered classification efforts; however, they were based on different approaches and lacked objective criteria for the differentiation of isolates. These differences have created discrepancies among systems, rendering discussions and comparisons across studies difficult. Although a system that used objective classification criteria was proposed by Diel and co-workers in 2012, the ample worldwide circulation and constant evolution of NDV, and utilization of only some of the criteria, led to identical naming and/or incorrect assigning of new sub/genotypes. To address these issues, an international consortium of experts was convened to undertake in-depth analyses of NDV genetic diversity. This consortium generated curated, up-to-date, complete fusion gene class I and class II datasets of all known NDV for public use, performed comprehensive phylogenetic neighbor-Joining, maximum-likelihood, Bayesian and nucleotide distance analyses, and compared these inference methods. An updated NDV classification and nomenclature system that incorporates phylogenetic topology, genetic distances, branch support, and epidemiological independence was developed. This new consensus system maintains two NDV classes and existing genotypes, identifies three new class II genotypes, and reduces the number of sub-genotypes. In order to track the ancestry of viruses, a dichotomous naming system for designating sub-genotypes was introduced. In addition, a pilot dataset and sub-trees rooting guidelines for rapid preliminary genotype identification of new isolates are provided. Guidelines for sequence dataset curation and phylogenetic inference, and a detailed comparison between the updated and previous systems are included. To increase the speed of phylogenetic inference and ensure consistency between laboratories, detailed guidelines for the use of a supercomputer are also provided. The proposed unified classification system will facilitate future studies of NDV evolution and epidemiology, and comparison of results obtained across the world.
Keywords: Avian paramyxovirus 1 (APMV-1); Classification; Genotype; Newcastle disease virus (NDV); Nomenclature; Phylogenetic analysis.
Crown Copyright © 2019. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Abolnik C., Mubamba C., Wandrag D.B.R., Horner R., Gummow B., Dautu G., Bisschop S.P.R. Tracing the origins of genotype VIIh Newcastle disease in southern Africa. Transbound. Emerg. Dis. 2018;65:e393–e403. - PubMed
-
- Ahmadi E., Pourbakhsh S.A., Ahmadi M., Mardani K., Talebi A. Phylogenetic characterization of virulent Newcastle disease viruses isolated during outbreaks in northwestern Iran in 2010. Arch. Virol. 2016;161:3151–3160. - PubMed
-
- Aldous E.W., Mynn J.K., Banks J., Alexander D.J. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003;32:239–256. - PubMed
-
- Aldous E.W., Fuller C.M., Mynn J.K., Alexander D.J. A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathol. 2004;33:258–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

