Single Nucleotide Polymorphisms of NUCB2 and their Genetic Associations with Milk Production Traits in Dairy Cows
- PMID: 31200542
- PMCID: PMC6627143
- DOI: 10.3390/genes10060449
Single Nucleotide Polymorphisms of NUCB2 and their Genetic Associations with Milk Production Traits in Dairy Cows
Abstract
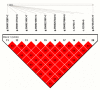
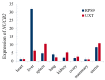
We previously used the RNA sequencing technique to detect the hepatic transcriptome of Chinese Holstein cows among the dry period, early lactation, and peak of lactation, and implied that the nucleobindin 2 (NUCB2) gene might be associated with milk production traits due to its expression being significantly increased in early lactation or peak of lactation as compared to dry period (q value < 0.05). Hence, in this study, we detected the single nucleotide polymorphisms (SNPs) of NUCB2 and analyzed their genetic associations with milk yield, fat yield, fat percentage, protein yield, and protein percentage. We re-sequenced the entire coding and 2000 bp of 5' and 3' flanking regions of NUCB2 by pooled sequencing, and identified ten SNPs, including one in 5' flanking region, two in 3' untranslated region (UTR), and seven in 3' flanking region. The single-SNP association analysis results showed that the ten SNPs were significantly associated with milk yield, fat yield, fat percentage, protein yield, or protein percentage in the first or second lactation (p values <= 1 × 10-4 and 0.05). In addition, we estimated the linkage disequilibrium (LD) of the ten SNPs by Haploview 4.2, and found that the SNPs were highly linked in one haplotype block (D' = 0.98-1.00), and the block was also significantly associated with at least one milk traits in the two lactations (p values: 0.0002-0.047). Further, we predicted the changes of transcription factor binding sites (TFBSs) that are caused by the SNPs in the 5' flanking region of NUCB2, and considered that g.35735477C>T might affect the expression of NUCB2 by changing the TFBSs for ETS transcription factor 3 (ELF3), caudal type homeobox 2 (CDX2), mammalian C-type LTR TATA box (VTATA), nuclear factor of activated T-cells (NFAT), and v-ets erythroblastosis virus E26 oncogene homolog (ERG) (matrix similarity threshold, MST > 0.85). However, the further study should be performed to verify the regulatory mechanisms of NUCB2 and its polymorphisms on milk traits. Our findings first revealed the genetic effects of NUCB2 on the milk traits in dairy cows, and suggested that the significant SNPs could be used in genomic selection to improve the accuracy of selection for dairy cattle breeding.
Keywords: SNP; TFBS; dairy cattle; genetic effect; milk yield and composition.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle.Genes (Basel). 2019 Apr 2;10(4):265. doi: 10.3390/genes10040265. Genes (Basel). 2019. PMID: 30986988 Free PMC article.
-
Single Nucleotide Polymorphisms of ALDH18A1 and MAT2A Genes and Their Genetic Associations with Milk Production Traits of Chinese Holstein Cows.Genes (Basel). 2022 Aug 12;13(8):1437. doi: 10.3390/genes13081437. Genes (Basel). 2022. PMID: 36011348 Free PMC article.
-
Identification of genetic associations and functional SNPs of bovine KLF6 gene on milk production traits in Chinese holstein.BMC Genom Data. 2023 Nov 28;24(1):72. doi: 10.1186/s12863-023-01175-w. BMC Genom Data. 2023. PMID: 38017423 Free PMC article.
-
Identification of single nucleotide polymorphisms of PIK3R1 and DUSP1 genes and their genetic associations with milk production traits in dairy cows.J Anim Sci Biotechnol. 2019 Nov 6;10:81. doi: 10.1186/s40104-019-0392-z. eCollection 2019. J Anim Sci Biotechnol. 2019. PMID: 31709048 Free PMC article.
-
Identification of Genetic Effects of ACADVL and IRF6 Genes with Milk Production Traits of Holstein Cattle in China.Genes (Basel). 2022 Dec 16;13(12):2393. doi: 10.3390/genes13122393. Genes (Basel). 2022. PMID: 36553659 Free PMC article.
Cited by
-
A Post-GWAS Functional Analysis Confirming Effects of Three BTA13 Genes CACNB2, SLC39A12, and ZEB1 on Dairy Cattle Reproduction.Front Genet. 2022 Jun 8;13:882951. doi: 10.3389/fgene.2022.882951. eCollection 2022. Front Genet. 2022. PMID: 35754833 Free PMC article.
-
Genomewide Association Analyses of Lactation Persistency and Milk Production Traits in Holstein Cattle Based on Imputed Whole-Genome Sequence Data.Genes (Basel). 2021 Nov 19;12(11):1830. doi: 10.3390/genes12111830. Genes (Basel). 2021. PMID: 34828436 Free PMC article.
-
A multi-tissue atlas of allelic-specific expression reveals the characteristics, mechanisms, and relationship with dominant effects in cattle.BMC Biol. 2025 May 30;23(1):148. doi: 10.1186/s12915-025-02257-0. BMC Biol. 2025. PMID: 40442638 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

