IL-23-producing IL-10Rα-deficient gut macrophages elicit an IL-22-driven proinflammatory epithelial cell response
- PMID: 31201258
- PMCID: PMC6697185
- DOI: 10.1126/sciimmunol.aau6571
IL-23-producing IL-10Rα-deficient gut macrophages elicit an IL-22-driven proinflammatory epithelial cell response
Abstract
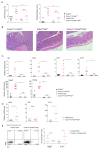
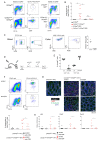
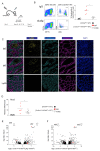
Cytokines maintain intestinal health, but precise intercellular communication networks remain poorly understood. Macrophages are immune sentinels of the intestinal tissue and are critical for gut homeostasis. Here, we show that in a murine inflammatory bowel disease (IBD) model based on macrophage-restricted interleukin-10 (IL-10) receptor deficiency (Cx3cr1Cre:Il10rafl/fl mice), proinflammatory mutant gut macrophages cause severe spontaneous colitis resembling the condition observed in children carrying IL-10R mutations. We establish macrophage-derived IL-23 as the driving factor of this pathology. Specifically, we report that Cx3cr1Cre:Il10rafl/fl:Il23afl/fl mice harboring macrophages deficient for both IL-10R and IL-23 are protected from colitis. By analyzing the epithelial response to proinflammatory macrophages, we provide evidence that T cells of colitic animals produce IL-22, which induces epithelial chemokine expression and detrimental neutrophil recruitment. Collectively, we define macrophage-specific contributions to the induction and pathogenesis of colitis, as manifested in mice harboring IL-10R deficiencies and human IBDs.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




References
-
- Joeris T, Müller-Luda K, Agace WW, Mowat AM. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. 2017;10:845–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

