An In Vitro Enrichment Strategy for Formulating Synergistic Synbiotics
- PMID: 31201276
- PMCID: PMC6677857
- DOI: 10.1128/AEM.01073-19
An In Vitro Enrichment Strategy for Formulating Synergistic Synbiotics
Abstract
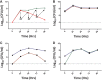
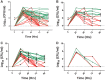
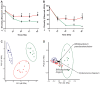
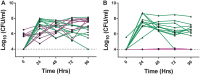
Research on the role of diet on gut and systemic health has led to considerable interest toward identifying novel therapeutic modulators of the gut microbiome, including the use of prebiotics and probiotics. However, various host responses have often been reported among many clinical trials. This is in part due to competitive exclusion as a result of the absence of ecological niches as well as host-mediated constraints via colonization resistance. In this research, we developed a novel in vitro enrichment (IVE) method for isolating autochthonous strains that can function as synergistic synbiotics and overcome these constraints. The method relied on stepwise in vitro fecal fermentations to enrich for and isolate Bifidobacterium strains that ferment the prebiotic xylooligosaccharide (XOS). We subsequently isolated Bifidobacterium longum subsp. longum CR15 and then tested its establishment in 20 unique fecal samples with or without XOS. The strain was established in up to 18 samples but only in the presence of XOS. Our findings revealed that the IVE method is suitable for isolating potential synergistic probiotic strains that possess the genetic and biochemical ability to ferment specific prebiotic substrates. The IVE method can be used as an initial high-throughput screen for probiotic selection and isolation prior to further characterization and in vivo tests.IMPORTANCE This study describes an in vitro enrichment method to formulate synergistic synbiotics that have potential for establishing autochthonous strains across multiple individuals. The rationale for this approach-that the chance of survival of a bacterial strain is improved by providing it with its required resources-is based on classic ecological theory. From these experiments, a human-derived strain, Bifidobacterium longum subsp. longum CR15, was identified as a xylooligosaccharide (XOS) fermenter in fecal environments and displayed synergistic effects in vitro The high rate of strain establishment observed in this study provides a basis for using synergistic synbiotics to overcome the responder/nonresponder phenomenon that occurs frequently in clinical trials with probiotic and prebiotic interventions. In addition, this approach can be applied in other protocols that require enrichment of specific bacterial populations prior to strain isolation.
Keywords: bifidobacteria; prebiotic; probiotic; synbiotic; xylooligosaccharide.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Evaluation of synbiotic combinations of commercial probiotic strains with different prebiotics in in vitro and ex vivo human gut microcosm model.Arch Microbiol. 2024 Jun 21;206(7):315. doi: 10.1007/s00203-024-04030-3. Arch Microbiol. 2024. PMID: 38904672
-
Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics.Microbiome. 2018 Jun 28;6(1):121. doi: 10.1186/s40168-018-0494-4. Microbiome. 2018. PMID: 29954454 Free PMC article. Clinical Trial.
-
In vitro fermentation of different fructo-oligosaccharides by Bifidobacterium strains for the selection of synbiotic combinations.Int J Food Microbiol. 2017 Feb 2;242:19-23. doi: 10.1016/j.ijfoodmicro.2016.11.011. Epub 2016 Nov 12. Int J Food Microbiol. 2017. PMID: 27866040
-
Effects of prebiotics, probiotics, and synbiotics on the infant gut microbiota and other health outcomes: A systematic review.Crit Rev Food Sci Nutr. 2023;63(22):5620-5642. doi: 10.1080/10408398.2021.2022595. Epub 2022 Jan 4. Crit Rev Food Sci Nutr. 2023. PMID: 37667870 Free PMC article. Review.
-
Ecological and molecular perspectives on responders and non-responders to probiotics and prebiotics.Curr Opin Biotechnol. 2022 Feb;73:108-120. doi: 10.1016/j.copbio.2021.06.023. Epub 2021 Aug 7. Curr Opin Biotechnol. 2022. PMID: 34375845 Review.
Cited by
-
A comprehensive review of synbiotics: an emerging paradigm in health promotion and disease management.World J Microbiol Biotechnol. 2024 Jul 27;40(9):280. doi: 10.1007/s11274-024-04085-w. World J Microbiol Biotechnol. 2024. PMID: 39060821 Review.
-
Biofilm-based delivery approaches and specific enrichment strategies of probiotics in the human gut.Gut Microbes. 2022 Jan-Dec;14(1):2126274. doi: 10.1080/19490976.2022.2126274. Gut Microbes. 2022. PMID: 36175161 Free PMC article. Review.
-
Inulin Fermentation by Lactobacilli and Bifidobacteria from Dairy Calves.Appl Environ Microbiol. 2020 Dec 17;87(1):e01738-20. doi: 10.1128/AEM.01738-20. Print 2020 Dec 17. Appl Environ Microbiol. 2020. PMID: 33008824 Free PMC article.
-
Megasphaera elsdenii, a commensal member of the gut microbiota, is associated with elevated gas production during in vitro fermentation.Gut Microbiome (Camb). 2023 Dec 21;5:e1. doi: 10.1017/gmb.2023.18. eCollection 2024. Gut Microbiome (Camb). 2023. PMID: 39290659 Free PMC article.
-
The Future of Synbiotics: Rational Formulation and Design.Front Microbiol. 2022 Jul 22;13:919725. doi: 10.3389/fmicb.2022.919725. eCollection 2022. Front Microbiol. 2022. PMID: 35935226 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

