Harnessing Human Microphysiology Systems as Key Experimental Models for Quantitative Systems Pharmacology
- PMID: 31201557
- PMCID: PMC6911651
- DOI: 10.1007/164_2019_239
Harnessing Human Microphysiology Systems as Key Experimental Models for Quantitative Systems Pharmacology
Abstract
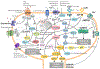
Two technologies that have emerged in the last decade offer a new paradigm for modern pharmacology, as well as drug discovery and development. Quantitative systems pharmacology (QSP) is a complementary approach to traditional, target-centric pharmacology and drug discovery and is based on an iterative application of computational and systems biology methods with multiscale experimental methods, both of which include models of ADME-Tox and disease. QSP has emerged as a new approach due to the low efficiency of success in developing therapeutics based on the existing target-centric paradigm. Likewise, human microphysiology systems (MPS) are experimental models complementary to existing animal models and are based on the use of human primary cells, adult stem cells, and/or induced pluripotent stem cells (iPSCs) to mimic human tissues and organ functions/structures involved in disease and ADME-Tox. Human MPS experimental models have been developed to address the relatively low concordance of human disease and ADME-Tox with engineered, experimental animal models of disease. The integration of the QSP paradigm with the use of human MPS has the potential to enhance the process of drug discovery and development.
Keywords: Computational models of ADME-Tox; Computational models of disease; DILI; Drug development; Drug discovery; Drug repurposing; Induced pluripotent stem cells; Microphysiology systems; Omics analyses; PBPK; Personalized medicine; Quantitative systems pharmacology; Toxicology.
Figures



Similar articles
-
ADME-Tox in drug discovery: integration of experimental and computational technologies.Drug Discov Today. 2003 Sep 15;8(18):852-61. doi: 10.1016/s1359-6446(03)02828-9. Drug Discov Today. 2003. PMID: 12963322 Review.
-
Integration not isolation: arguing the case for quantitative and systems pharmacology in drug discovery and development.Drug Discov Today. 2011 Dec;16(23-24):1031-6. doi: 10.1016/j.drudis.2011.10.001. Epub 2011 Oct 13. Drug Discov Today. 2011. PMID: 22020181 Review.
-
Using quantitative systems pharmacology for novel drug discovery.Expert Opin Drug Discov. 2015 Dec;10(12):1315-31. doi: 10.1517/17460441.2015.1082543. Epub 2015 Aug 25. Expert Opin Drug Discov. 2015. PMID: 26328768 Review.
-
A Perspective on Implementing a Quantitative Systems Pharmacology Platform for Drug Discovery and the Advancement of Personalized Medicine.J Biomol Screen. 2016 Jul;21(6):521-34. doi: 10.1177/1087057116635818. Epub 2016 Mar 8. J Biomol Screen. 2016. PMID: 26962875 Free PMC article.
-
Evolution of Experimental Models of the Liver to Predict Human Drug Hepatotoxicity and Efficacy.Clin Liver Dis. 2017 Feb;21(1):197-214. doi: 10.1016/j.cld.2016.08.013. Clin Liver Dis. 2017. PMID: 27842772 Free PMC article. Review.
Cited by
-
Organ-On-Chip Technology: The Future of Feto-Maternal Interface Research?Front Physiol. 2020 Jun 30;11:715. doi: 10.3389/fphys.2020.00715. eCollection 2020. Front Physiol. 2020. PMID: 32695021 Free PMC article. Review.
-
Analysis of reproducibility and robustness of OrganoPlate® 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs.Toxicol In Vitro. 2022 Dec;85:105464. doi: 10.1016/j.tiv.2022.105464. Epub 2022 Aug 31. Toxicol In Vitro. 2022. PMID: 36057418 Free PMC article.
-
Human biomimetic liver microphysiology systems in drug development and precision medicine.Nat Rev Gastroenterol Hepatol. 2021 Apr;18(4):252-268. doi: 10.1038/s41575-020-00386-1. Epub 2020 Dec 17. Nat Rev Gastroenterol Hepatol. 2021. PMID: 33335282 Free PMC article. Review.
-
The Combination of a Human Biomimetic Liver Microphysiology System with BIOLOGXsym, a Quantitative Systems Toxicology (QST) Modeling Platform for Macromolecules, Provides Mechanistic Understanding of Tocilizumab- and GGF2-Induced Liver Injury.Int J Mol Sci. 2023 Jun 2;24(11):9692. doi: 10.3390/ijms24119692. Int J Mol Sci. 2023. PMID: 37298645 Free PMC article.
-
The Microbiome and the Gut-Liver-Brain Axis for Central Nervous System Clinical Pharmacology: Challenges in Specifying and Integrating In Vitro and In Silico Models.Clin Pharmacol Ther. 2020 Nov;108(5):929-948. doi: 10.1002/cpt.1870. Epub 2020 May 29. Clin Pharmacol Ther. 2020. PMID: 32347548 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

