Altered Levels of Proteins and Phosphoproteins, in the Absence of Early Causative Transcriptional Changes, Shape the Molecular Pathogenesis in the Brain of Young Presymptomatic Ki91 SCA3/MJD Mouse
- PMID: 31201651
- PMCID: PMC6834541
- DOI: 10.1007/s12035-019-01643-4
Altered Levels of Proteins and Phosphoproteins, in the Absence of Early Causative Transcriptional Changes, Shape the Molecular Pathogenesis in the Brain of Young Presymptomatic Ki91 SCA3/MJD Mouse
Abstract
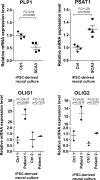
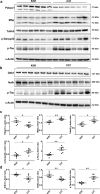
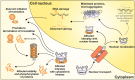
Spinocerebellar ataxia type 3 (SCA3/MJD) is a polyQ neurodegenerative disease where the presymptomatic phase of pathogenesis is unknown. Therefore, we investigated the molecular network of transcriptomic and proteomic triggers in young presymptomatic SCA3/MJD brain from Ki91 knock-in mouse. We found that transcriptional dysregulations resulting from mutant ataxin-3 are not occurring in young Ki91 mice, while old Ki91 mice and also postmitotic patient SCA3 neurons demonstrate the late transcriptomic changes. Unlike the lack of early mRNA changes, we have identified numerous early changes of total proteins and phosphoproteins in 2-month-old Ki91 mouse cortex and cerebellum. We discovered the network of processes in presymptomatic SCA3 with three main groups of disturbed processes comprising altered proteins: (I) modulation of protein levels and DNA damage (Pabpc1, Ddb1, Nedd8), (II) formation of neuronal cellular structures (Tubb3, Nefh, p-Tau), and (III) neuronal function affected by processes following perturbed cytoskeletal formation (Mt-Co3, Stx1b, p-Syn1). Phosphoproteins downregulate in the young Ki91 mouse brain and their phosphosites are associated with kinases that interact with ATXN3 such as casein kinase, Camk2, and kinases controlled by another Atxn3 interactor p21 such as Gsk3, Pka, and Cdk kinases. We conclude that the onset of SCA3 pathology occurs without altered transcript level and is characterized by changed levels of proteins responsible for termination of translation, DNA damage, spliceosome, and protein phosphorylation. This disturbs global cellular processes such as cytoskeleton and transport of vesicles and mitochondria along axons causing energy deficit and neurodegeneration also manifesting in an altered level of transcripts at later ages.
Keywords: Ataxia; Ataxin-3; CAG; Knock-in; MJD; Mouse; Phosphoproteome; PolyQ; Presymptomatic; Proteome; SCA3; Spinocerebellar.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
A new humanized ataxin-3 knock-in mouse model combines the genetic features, pathogenesis of neurons and glia and late disease onset of SCA3/MJD.Neurobiol Dis. 2015 Jan;73:174-88. doi: 10.1016/j.nbd.2014.09.020. Epub 2014 Oct 7. Neurobiol Dis. 2015. PMID: 25301414
-
Broad Influence of Mutant Ataxin-3 on the Proteome of the Adult Brain, Young Neurons, and Axons Reveals Central Molecular Processes and Biomarkers in SCA3/MJD Using Knock-In Mouse Model.Front Mol Neurosci. 2021 Jun 17;14:658339. doi: 10.3389/fnmol.2021.658339. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34220448 Free PMC article.
-
Comparison of spinocerebellar ataxia type 3 mouse models identifies early gain-of-function, cell-autonomous transcriptional changes in oligodendrocytes.Hum Mol Genet. 2017 Sep 1;26(17):3362-3374. doi: 10.1093/hmg/ddx224. Hum Mol Genet. 2017. PMID: 28854700 Free PMC article.
-
Autophagy in Spinocerebellar Ataxia Type 3: From Pathogenesis to Therapeutics.Int J Mol Sci. 2023 Apr 17;24(8):7405. doi: 10.3390/ijms24087405. Int J Mol Sci. 2023. PMID: 37108570 Free PMC article. Review.
-
Planning Future Clinical Trials for Machado-Joseph Disease.Adv Exp Med Biol. 2018;1049:321-348. doi: 10.1007/978-3-319-71779-1_17. Adv Exp Med Biol. 2018. PMID: 29427112 Review.
Cited by
-
From Pathogenesis to Novel Therapeutics for Spinocerebellar Ataxia Type 3: Evading Potholes on the Way to Translation.Neurotherapeutics. 2019 Oct;16(4):1009-1031. doi: 10.1007/s13311-019-00798-1. Neurotherapeutics. 2019. PMID: 31691128 Free PMC article. Review.
-
Plasma PolyQ-ATXN3 Levels Associate With Cerebellar Degeneration and Behavioral Abnormalities in a New AAV-Based SCA3 Mouse Model.Front Cell Dev Biol. 2022 Mar 21;10:863089. doi: 10.3389/fcell.2022.863089. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35386195 Free PMC article.
-
A Novel SCA3 Knock-in Mouse Model Mimics the Human SCA3 Disease Phenotype Including Neuropathological, Behavioral, and Transcriptional Abnormalities Especially in Oligodendrocytes.Mol Neurobiol. 2022 Jan;59(1):495-522. doi: 10.1007/s12035-021-02610-8. Epub 2021 Oct 30. Mol Neurobiol. 2022. PMID: 34716557 Free PMC article.
-
Tau and neurofilament light-chain as fluid biomarkers in spinocerebellar ataxia type 3.Eur J Neurol. 2022 Aug;29(8):2439-2452. doi: 10.1111/ene.15373. Epub 2022 May 26. Eur J Neurol. 2022. PMID: 35478426 Free PMC article.
-
The Josephin domain (JD) containing proteins are predicted to bind to the same interactors: Implications for spinocerebellar ataxia type 3 (SCA3) studies using Drosophila melanogaster mutants.Front Mol Neurosci. 2023 Mar 15;16:1140719. doi: 10.3389/fnmol.2023.1140719. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37008788 Free PMC article.
References
-
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. - DOI - PMC - PubMed
-
- Todi SV, Scaglione KM, Blount JR, Basrur V, Conlon KP, Pastore A, Elenitoba-Johnson K, Paulson HL. Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J Biol Chem. 2010;285:39303–39313. doi: 10.1074/jbc.M110.181610. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

