mCerulean3-Based Cameleon Sensor to Explore Mitochondrial Ca2+ Dynamics In Vivo
- PMID: 31203189
- PMCID: PMC6581653
- DOI: 10.1016/j.isci.2019.05.031
mCerulean3-Based Cameleon Sensor to Explore Mitochondrial Ca2+ Dynamics In Vivo
Erratum in
-
mCerulean3-Based Cameleon Sensor to Explore Mitochondrial Ca2+ Dynamics In Vivo.iScience. 2019 Sep 27;19:161. doi: 10.1016/j.isci.2019.07.031. Epub 2019 Jul 30. iScience. 2019. PMID: 31374427 Free PMC article. No abstract available.
Abstract
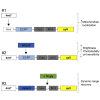
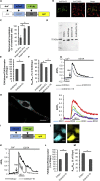
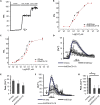
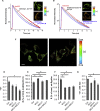
Genetically Encoded Ca2+ Indicators (GECIs) are extensively used to study organelle Ca2+ homeostasis, although some available probes are still plagued by a number of problems, e.g., low fluorescence intensity, partial mistargeting, and pH sensitivity. Furthermore, in the most commonly used mitochondrial Förster Resonance Energy Transfer based-GECIs, the donor protein ECFP is characterized by a double exponential lifetime that complicates the fluorescence lifetime analysis. We have modified the cytosolic and mitochondria-targeted Cameleon GECIs by (1) substituting the donor ECFP with mCerulean3, a brighter and more stable fluorescent protein with a single exponential lifetime; (2) extensively modifying the constructs to improve targeting efficiency and fluorescence changes caused by Ca2+ binding; and (3) inserting the cDNAs into adeno-associated viral vectors for in vivo expression. The probes have been thoroughly characterized in situ by fluorescence microscopy and Fluorescence Lifetime Imaging Microscopy, and examples of their ex vivo and in vivo applications are described.
Keywords: Biological Sciences Tools; Cell Biology; Optical Imaging.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A New Transgenic Mouse Line for Imaging Mitochondrial Calcium Signals.Function (Oxf). 2021 Feb 25;2(3):zqab012. doi: 10.1093/function/zqab012. eCollection 2021. Function (Oxf). 2021. PMID: 35330679 Free PMC article.
-
Generation and Characterization of a New FRET-Based Ca2+ Sensor Targeted to the Nucleus.Int J Mol Sci. 2021 Sep 14;22(18):9945. doi: 10.3390/ijms22189945. Int J Mol Sci. 2021. PMID: 34576104 Free PMC article.
-
Live Mitochondrial or Cytosolic Calcium Imaging Using Genetically-encoded Cameleon Indicator in Mammalian Cells.Bio Protoc. 2020 Feb 5;10(3):e3504. doi: 10.21769/BioProtoc.3504. eCollection 2020 Feb 5. Bio Protoc. 2020. PMID: 33654731 Free PMC article.
-
Newly engineered cyan fluorescent proteins with enhanced performances for live cell FRET imaging.Biotechnol J. 2014 Feb;9(2):180-91. doi: 10.1002/biot.201300198. Epub 2013 Dec 19. Biotechnol J. 2014. PMID: 24357633 Review.
-
New red-fluorescent calcium indicators for optogenetics, photoactivation and multi-color imaging.Biochim Biophys Acta. 2014 Oct;1843(10):2284-306. doi: 10.1016/j.bbamcr.2014.03.010. Epub 2014 Mar 27. Biochim Biophys Acta. 2014. PMID: 24681159 Review.
Cited by
-
Calcium Signaling and Mitochondrial Function in Presenilin 2 Knock-Out Mice: Looking for Any Loss-of-Function Phenotype Related to Alzheimer's Disease.Cells. 2021 Jan 21;10(2):204. doi: 10.3390/cells10020204. Cells. 2021. PMID: 33494218 Free PMC article.
-
Advanced Imaging Techniques for the Characterization of Subcellular Organelle Structure in Pancreatic Islet β Cells.Compr Physiol. 2023 Dec 29;14(1):5243-5267. doi: 10.1002/cphy.c230002. Compr Physiol. 2023. PMID: 38158370 Free PMC article.
-
Molecular Spies in Action: Genetically Encoded Fluorescent Biosensors Light up Cellular Signals.Chem Rev. 2024 Nov 27;124(22):12573-12660. doi: 10.1021/acs.chemrev.4c00293. Epub 2024 Nov 13. Chem Rev. 2024. PMID: 39535501 Free PMC article. Review.
-
Probing spatiotemporally organized GPCR signaling using genetically encoded molecular tools.Exp Mol Med. 2025 Jul;57(7):1432-1442. doi: 10.1038/s12276-025-01485-2. Epub 2025 Jul 1. Exp Mol Med. 2025. PMID: 40588530 Free PMC article. Review.
-
Infection-induced peripheral mitochondria fission drives ER encapsulations and inter-mitochondria contacts that rescue bioenergetics.Nat Commun. 2024 Aug 27;15(1):7352. doi: 10.1038/s41467-024-51680-4. Nat Commun. 2024. PMID: 39187492 Free PMC article.
References
-
- Arnaudeau S., Kelley W.L., Walsh J.V., Jr., Demaurex N. Mitochondria recycle Ca2+ to the endoplasmic reticulum and prevent the depletion of neighboring endoplasmic reticulum regions. J. Biol. Chem. 2001;276:29430–29439. - PubMed
- Arnaudeau S., W.L. Kelley, J.V. Walsh Jr, N. Demaurex(2001). Mitochondria recycle Ca2+ to the endoplasmic reticulum and prevent the depletion of neighboring endoplasmic reticulum regions J. Biol. Chem., 276, pp. 29430-29439. - PubMed
-
- Bish L.T., Morine K., Sleeper M.M., Sanmiguel J., Wu D., Gao G.P., Wilson J.M., Sweeney H.L. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum. Gene Ther. 2008;19:1359–1368. - PMC - PubMed
- Bish, L.T., Morine, K., Sleeper, M.M., Sanmiguel, J., Wu, D., Gao, G.P., Wilson, J.M., and Sweeney, H.L.. (2008). Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum. Gene Ther. 19, 1359-1368. - PMC - PubMed
-
- Borst J.W., Laptenok S.P., Westphal A.H., Kuhnemuth R., Hornen H., Visser N.V., Kalinin S., Aker J., van Hoek A., Seidel C.A. Structural changes of yellow Cameleon domains observed by quantitative FRET analysis and polarized fluorescence correlation spectroscopy. Biophys. J. 2008;95:5399–5411. - PMC - PubMed
- Borst, J.W., Laptenok, S.P., Westphal, A.H., Kuhnemuth, R., Hornen, H., Visser, N.V., Kalinin, S., Aker, J., van Hoek, A., Seidel, C.A., et al. (2008). Structural changes of yellow Cameleon domains observed by quantitative FRET analysis and polarized fluorescence correlation spectroscopy. Biophys. J. 95, 5399-5411. - PMC - PubMed
-
- Cárdenas C., Miller R.A., Smith I., Bui T., Molgó J., Müller M., Vais H., Cheung K.H., Yang J., Parker I. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. - PMC - PubMed
- Cardenas C., Miller R.A., Smith I., Bui T., Molgo J., Muller M., Vais H., Cheung K.H., Yang J., Parker I., et al.(2010) Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria.Cell. ;142:270-283. - PMC - PubMed
-
- Chalmers S., Nicholls D.G. The relationship between free and total calcium concentrations in the matrix of liver and brain mitochondria. J. Biol. Chem. 2003;278:19062–19070. - PubMed
- Chalmers S., Nicholls D.G.. (2003) The relationship between free and total calcium concentrations in the matrix of liver and brain mitochondria. J. Biol. Chem.. 278:19062-19070. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

