Genes for Good: Engaging the Public in Genetics Research via Social Media
- PMID: 31204010
- PMCID: PMC6612519
- DOI: 10.1016/j.ajhg.2019.05.006
Genes for Good: Engaging the Public in Genetics Research via Social Media
Erratum in
-
Genes for Good: Engaging the Public in Genetics Research via Social Media.Am J Hum Genet. 2019 Aug 1;105(2):441-442. doi: 10.1016/j.ajhg.2019.07.009. Am J Hum Genet. 2019. PMID: 31374205 Free PMC article. No abstract available.
Abstract
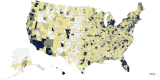
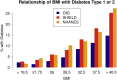
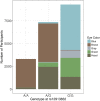
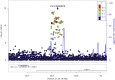
The Genes for Good study uses social media to engage a large, diverse participant pool in genetics research and education. Health history and daily tracking surveys are administered through a Facebook application, and participants who complete a minimum number of surveys are mailed a saliva sample kit ("spit kit") to collect DNA for genotyping. As of March 2019, we engaged >80,000 individuals, sent spit kits to >32,000 individuals who met minimum participation requirements, and collected >27,000 spit kits. Participants come from all 50 states and include a diversity of ancestral backgrounds. Rates of important chronic health indicators are consistent with those estimated for the general U.S. population using more traditional study designs. However, our sample is younger and contains a greater percentage of females than the general population. As one means of verifying data quality, we have replicated genome-wide association studies (GWASs) for exemplar traits, such as asthma, diabetes, body mass index (BMI), and pigmentation. The flexible framework of the web application makes it relatively simple to add new questionnaires and for other researchers to collaborate. We anticipate that the study sample will continue to grow and that future analyses may further capitalize on the strengths of the longitudinal data in combination with genetic information.
Keywords: asthma; body mass index; complex traits; diabetes; direct to participant research; genome-wide association study; genotyping array; participant engagement; population study; social media.
Copyright © 2019 American Society of Human Genetics. All rights reserved.
Figures



References
-
- Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. - PMC - PubMed
- Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., Klemm, A., Flicek, P., Manolio, T., Hindorff, L., and Parkinson, H. (2014). The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001-D1006. - PMC - PubMed
-
- Stoeklé H.C., Mamzer-Bruneel M.F., Vogt G., Hervé C. 23andMe: a new two-sided data-banking market model. BMC Med. Ethics. 2016;17:19. - PMC - PubMed
- Stoekle, H.C., Mamzer-Bruneel, M.F., Vogt, G., and Herve, C. (2016). 23andMe: a new two-sided data-banking market model. BMC Med. Ethics 17, 19. - PMC - PubMed
-
- Royal C.D., Novembre J., Fullerton S.M., Goldstein D.B., Long J.C., Bamshad M.J., Clark A.G. Inferring genetic ancestry: opportunities, challenges, and implications. Am. J. Hum. Genet. 2010;86:661–673. - PMC - PubMed
- Royal, C.D., Novembre, J., Fullerton, S.M., Goldstein, D.B., Long, J.C., Bamshad, M.J., and Clark, A.G. (2010). Inferring genetic ancestry: opportunities, challenges, and implications. Am. J. Hum. Genet. 86, 661-673. - PMC - PubMed
-
- Agurs-Collins T., Ferrer R., Ottenbacher A., Waters E.A., O’Connell M.E., Hamilton J.G. Public Awareness of Direct-to-Consumer Genetic Tests: Findings from the 2013 U.S. Health Information National Trends Survey. J. Cancer Educ. 2015;30:799–807. - PMC - PubMed
- Agurs-Collins, T., Ferrer, R., Ottenbacher, A., Waters, E.A., O’Connell, M.E., and Hamilton, J.G. (2015). Public Awareness of Direct-to-Consumer Genetic Tests: Findings from the 2013 U.S. Health Information National Trends Survey. J. Cancer Educ. 30, 799-807. - PMC - PubMed
-
- Pedersen E.R., Kurz J. Using Facebook for Health-related Research Study Recruitment and Program Delivery. Curr Opin Psychol. 2016;9:38–43. - PMC - PubMed
- Pedersen, E.R., and Kurz, J. (2016). Using Facebook for Health-related Research Study Recruitment and Program Delivery. Curr Opin Psychol 9, 38-43. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

