Mechanical Function of the Nucleus in Force Generation during Epithelial Morphogenesis
- PMID: 31204174
- PMCID: PMC6658619
- DOI: 10.1016/j.devcel.2019.05.027
Mechanical Function of the Nucleus in Force Generation during Epithelial Morphogenesis
Abstract
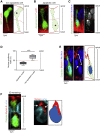
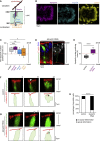
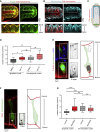
Mechanical forces are critical regulators of cell shape changes and developmental morphogenetic processes. Forces generated along the epithelium apico-basal cell axis have recently emerged as essential for tissue remodeling in three dimensions. Yet the cellular machinery underlying those orthogonal forces remains poorly described. We found that during Drosophila leg folding cells eventually committed to die produce apico-basal forces through the formation of a dynamic actomyosin contractile tether connecting the apical surface to a basally relocalized nucleus. We show that the nucleus is anchored to basal adhesions by a basal F-actin network and constitutes an essential component of the force-producing machinery. Finally, we demonstrate force transmission to the apical surface and the basal nucleus by laser ablation. Thus, this work reveals that the nucleus, in addition to its role in genome protection, actively participates in mechanical force production and connects the contractile actomyosin cytoskeleton to basal adhesions.
Keywords: Drosophila; LINC; Live imaging; Talin; actomyosin; apoptosis; basal adhesion; linker of nucleoskeleton and cytoskeleton complex; mechanical forces; morphogenesis; nucleus.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

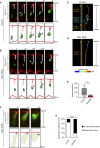

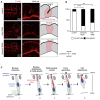
Comment in
-
Pulling the Roof Down on Anchored Nuclei.Dev Cell. 2019 Jul 22;50(2):130-131. doi: 10.1016/j.devcel.2019.07.003. Dev Cell. 2019. PMID: 31336096
References
-
- Ambrosini A., Gracia M., Proag A., Rayer M., Monier B., Suzanne M. Apoptotic forces in tissue morphogenesis. Mech. Dev. 2017;144:33–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

