WDR45 contributes to neurodegeneration through regulation of ER homeostasis and neuronal death
- PMID: 31204559
- PMCID: PMC6999610
- DOI: 10.1080/15548627.2019.1630224
WDR45 contributes to neurodegeneration through regulation of ER homeostasis and neuronal death
Abstract
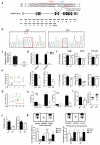
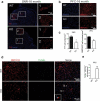
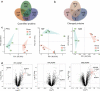
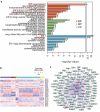
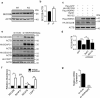
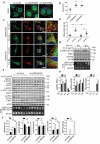
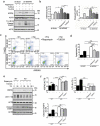
Mutations in the macroautophagy/autophagy gene WDR45 cause β-propeller protein-associated neurodegeneration (BPAN); however the molecular and cellular mechanism of the disease process is largely unknown. Here we generated constitutive wdr45 knockout (KO) mice that displayed cognitive impairments, abnormal synaptic transmission and lesions in several brain regions. Immunohistochemistry analysis showed loss of neurons in prefrontal cortex and basal ganglion in aged mice, and increased apoptosis in prefrontal cortex, recapitulating a hallmark of neurodegeneration. Quantitative proteomic analysis showed accumulation of endoplasmic reticulum (ER) proteins in KO mouse. At the cellular level, accumulation of ER proteins due to WDR45 deficiency resulted in increased ER stress and impaired ER quality control. The unfolded protein response (UPR) was elevated through ERN1/IRE1 or EIF2AK3/PERK pathway, and eventually led to neuronal apoptosis. Suppression of ER stress or activation of autophagy through MTOR inhibition alleviated cell death. Thus, the loss of WDR45 cripples macroautophagy machinery in neurons and leads to impairment in organelle autophagy, which provides a mechanistic understanding of cause of BPAN and a potential therapeutic strategy to treat this genetic disorder.Abbreviations: 7-ADD: 7-aminoactinomycin D; ASD: autistic spectrum disorder; ATF6: activating transcription factor 6; ATG: autophagy-related; BafA1: bafilomycin A1; BCAP31: B cell receptor associated protein 31; BPAN: β-propeller protein-associated neurodegeneration; CCCP: carbonyl cyanide m-chlorophenylhydrazone; CDIPT: CDP-diacylglycerol-inositol 3-phosphatidyltransferase (phosphatidylinositol synthase); DDIT3/CHOP: DNA-damage inducible transcript 3; EIF2A: eukaryotic translation initiation factor 2A; EIF2AK3/PERK: eukaryotic translation initiation factor 2 alpha kinase 3; ER: endoplasmic reticulum; ERN1/IRE1: endoplasmic reticulum to nucleus signaling 1; GFP: green fluorescent protein; HIP: hippocampus; HSPA5/GRP78: heat shock protein family A (HSP70) member 5; KO: knockout; LAMP1: lysosomal-associated membrane 1; mEPSCs: miniature excitatory postsynaptic currents; MG132: N-benzyloxycarbonyl-L-leucyl-L-leucyl-L-leucinal; MIB: mid-brain; MTOR: mechanistic target of rapamycin kinase; PCR: polymerase chain reaction; PFA: paraformaldehyde; PFC: prefrontal cortex; PRM: parallel reaction monitoring; RBFOX3/NEUN: RNA binding protein, fox-1 homolog [C. elegans] 3; RTN3: reticulon 3; SEC22B: SEC22 homolog B, vesicle trafficking protein; SEC61B: SEC61 translocon beta subunit; SEM: standard error of the mean; SNR: substantia nigra; SQSTM1/p62: sequestosome 1; TH: tyrosine hydroxylase; Tm: tunicamycin; TMT: tandem mass tag; TUDCA: tauroursodeoxycholic acid; TUNEL: terminal deoxynucleotidyl transferase dUTP nick-end labeling; UPR: unfolded protein response; WDR45: WD repeat domain 45; WT: wild type; XBP1: X-box binding protein 1.
Keywords: ER stress; UPR; WDR45; neuronal apoptosis; quantitative proteomics.
Figures

References
-
- Choi AMK, Ryter SW, Levine B.. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. - PubMed
-
- Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006. 04/19/online;441:885. - PubMed
-
- Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006. 04/19/online;441:880. - PubMed
-
- Menzies FM, Fleming A, Caricasole A, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015–1034. - PubMed
-
- Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases [Review Article]. Nat Rev Neurosci. 2015. 05/20/online;16:345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
