Absolute quantification of cohesin, CTCF and their regulators in human cells
- PMID: 31204999
- PMCID: PMC6606026
- DOI: 10.7554/eLife.46269
Absolute quantification of cohesin, CTCF and their regulators in human cells
Abstract
The organisation of mammalian genomes into loops and topologically associating domains (TADs) contributes to chromatin structure, gene expression and recombination. TADs and many loops are formed by cohesin and positioned by CTCF. In proliferating cells, cohesin also mediates sister chromatid cohesion, which is essential for chromosome segregation. Current models of chromatin folding and cohesion are based on assumptions of how many cohesin and CTCF molecules organise the genome. Here we have measured absolute copy numbers and dynamics of cohesin, CTCF, NIPBL, WAPL and sororin by mass spectrometry, fluorescence-correlation spectroscopy and fluorescence recovery after photobleaching in HeLa cells. In G1-phase, there are ~250,000 nuclear cohesin complexes, of which ~ 160,000 are chromatin-bound. Comparison with chromatin immunoprecipitation-sequencing data implies that some genomic cohesin and CTCF enrichment sites are unoccupied in single cells at any one time. We discuss the implications of these findings for how cohesin can contribute to genome organisation and cohesion.
Keywords: CTCF; cell biology; chromosomes; cohesin; fluorescence-correlation spectroscopy; gene expression; genome organization; human; mass spectrometry; sister chromatid cohesion.
© 2019, Holzmann et al.
Conflict of interest statement
JH, AP, KN, MH, NW, BK, JF, GD, WT, RL, RS, GB, BN, KM, ID, JE, JP No competing interests declared
Figures
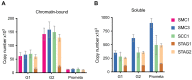
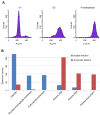

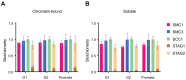
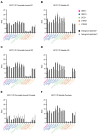



Comment in
-
Determining cellular CTCF and cohesin abundances to constrain 3D genome models.Elife. 2019 Jun 17;8:e40164. doi: 10.7554/eLife.40164. Elife. 2019. PMID: 31205001 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- WWTF LS09-13/Vienna Science and Technology Fund/International
- International PhD Programme/European Molecular Biology Laboratory/International
- U01 DA047728/DA/NIDA NIH HHS/United States
- Laura Bassi Centre for Optimized Structural Studies grant FFG-840283/Austrian Research Promotion Agency/International
- Common Fund 4D Nucleome Program (U01 DA047728)/NH/NIH HHS/United States
- Allen Distinguished Investigator Program/Paul G. Allen Frontiers Group/International
- I 3686/FWF_/Austrian Science Fund FWF/Austria
- Common Fund 4D Nucleome Program (U01 EB021223)/NH/NIH HHS/United States
- ALTF 1335-2016/European Molecular Biology Organization/International
- Common Fund 4D Nucleome Program (U01 EB021223 / U01 DA047728)/NH/NIH HHS/United States
- 693949/Horizon 2020 Framework Programme/International
- LT001527/2017/Human Frontier Science Program/International
- I 3686-B25 MEIOREC - ERA-CAPS/Austrian Science Fund/International
- 823839/Horizon 2020 Framework Programme/International
- 503464 (MitoCheck)/Sixth Framework Programme/International
- 653706/Horizon 2020 Framework Programme/International
- U01 EB021223/EB/NIBIB NIH HHS/United States
- Wittgenstein award Z196-B20/Austrian Science Fund/International
- 241548 (MitoSys)/Seventh Framework Programme/International
- FFG-834223/Austrian Research Promotion Agency/International
- FWF special research program SFB F34/Austrian Science Fund/International
- FFG-852936/Austrian Research Promotion Agency/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

