Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviours of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway
- PMID: 31207033
- PMCID: PMC6653555
- DOI: 10.1111/jcmm.14362
Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviours of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway
Abstract
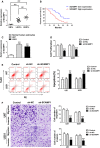
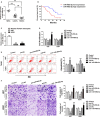
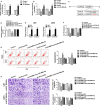
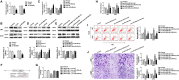
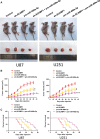
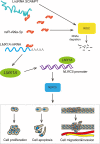
Dysregulation of long non-coding RNAs (lncRNAs) confirm that it plays a crucial role in tumourigenesis and malignant progression of glioma. The present study demonstrated that LncRNA secretory carrier membrane protein 1 (SCAMP1) was up-regulated and functioned as an oncogene in glioma cells. In addition, miR-499a-5p was down-regulated meanwhile exerted tumour-suppressive function in glioma cells. Subsequently, inhibition of SCAMP1 significantly restrained the cell proliferation, migration and invasion, as well as promoted apoptosis by acting as a molecular sponge of miR-499a-5p. Transcription factor LIM homeobox transcription factor 1, alpha (LMX1A) was overexpressed in glioma tissues and cells. Moreover, miR-499a-5p targeted LMX1A 3'-UTR in a sequence-specific manner. Hence, down-regulation of SCAMP1 remarkably reduced the expression level of LMX1A, indicating that LMX1A participated in miR-499a-5p-induced tumour-suppressive effects on glioma cells. Furthermore, knockdown of LMX1A decreased NLR family, CARD domain containing 5 (NLRC5) mRNA and protein expression levels through directly binding to the NLRC5 promoter region. Down-regulation of NLRC5 obviously inhibited malignant biological behaviours of glioma cells through attenuating the activity of Wnt/β-catenin signalling pathway. In conclusion, our study clarifies that SCAMP1/miR-499a-5p/LMX1A/NLRC5 axis plays a critical role in modulating malignant progression of glioma cells, which provide a novel therapeutic strategy for glioma treatment.
Keywords: LMX1A; NLRC5; SCAMP1; glioma; long non-coding RNAs; miR-499a-5p.
© 2019 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
ceRNA network of lncRNA MIR210HG/miR-377-3p/LMX1A in malignant proliferation of glioma cells.Genes Genomics. 2022 Dec;44(12):1445-1455. doi: 10.1007/s13258-022-01312-2. Epub 2022 Oct 5. Genes Genomics. 2022. PMID: 36197580
-
An upstream open reading frame regulates vasculogenic mimicry of glioma via ZNRD1-AS1/miR-499a-5p/ELF1/EMI1 pathway.J Cell Mol Med. 2020 Jun;24(11):6120-6136. doi: 10.1111/jcmm.15217. Epub 2020 May 5. J Cell Mol Med. 2020. PMID: 32368853 Free PMC article.
-
m6A-Mediated Upregulation of lncRNA CHASERR Promotes the Progression of Glioma by Modulating the miR-6893-3p/TRIM14 Axis.Mol Neurobiol. 2024 Aug;61(8):5418-5440. doi: 10.1007/s12035-023-03911-w. Epub 2024 Jan 9. Mol Neurobiol. 2024. PMID: 38193984
-
Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma.Mol Cell Biochem. 2021 Jun;476(6):2317-2335. doi: 10.1007/s11010-021-04080-x. Epub 2021 Feb 13. Mol Cell Biochem. 2021. PMID: 33582947 Review.
-
Current advances of long non-coding RNAs mediated by wnt signaling in glioma.Pathol Res Pract. 2020 Aug;216(8):153008. doi: 10.1016/j.prp.2020.153008. Epub 2020 May 25. Pathol Res Pract. 2020. PMID: 32703485 Review.
Cited by
-
The up-regulation of LRIG1 expression inhibits the proliferation, apoptosis and invasion of glioma cells.Am J Transl Res. 2022 Feb 15;14(2):788-797. eCollection 2022. Am J Transl Res. 2022. PMID: 35273685 Free PMC article.
-
Circular RNA circ_0079593 facilitates glioma development via modulating miR-324-5p/XBP1 axis.Metab Brain Dis. 2022 Oct;37(7):2389-2403. doi: 10.1007/s11011-022-01040-2. Epub 2022 Jul 6. Metab Brain Dis. 2022. PMID: 35793013
-
LncRNA TMPO-AS1 Promotes Proliferation and Invasion by Sponging miR-383-5p in Glioma Cells.Cancer Manag Res. 2020 Nov 23;12:12001-12009. doi: 10.2147/CMAR.S282539. eCollection 2020. Cancer Manag Res. 2020. PMID: 33262650 Free PMC article.
-
Linc00475 promotes the progression of glioma by regulating the miR-141-3p/YAP1 axis.J Cell Mol Med. 2021 Jan;25(1):463-472. doi: 10.1111/jcmm.16100. Epub 2020 Dec 18. J Cell Mol Med. 2021. PMID: 33336871 Free PMC article.
-
ceRNA network of lncRNA MIR210HG/miR-377-3p/LMX1A in malignant proliferation of glioma cells.Genes Genomics. 2022 Dec;44(12):1445-1455. doi: 10.1007/s13258-022-01312-2. Epub 2022 Oct 5. Genes Genomics. 2022. PMID: 36197580
References
-
- Spinelli GP, Miele E, Lo Russo G, et al. Chemotherapy and target therapy in the management of adult high‐grade gliomas. Curr Cancer Drug Targets. 2012;12:1016‐1031. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

