Nervous-Like Circuits in the Ribosome Facts, Hypotheses and Perspectives
- PMID: 31207893
- PMCID: PMC6627100
- DOI: 10.3390/ijms20122911
Nervous-Like Circuits in the Ribosome Facts, Hypotheses and Perspectives
Abstract
In the past few decades, studies on translation have converged towards the metaphor of a "ribosome nanomachine"; they also revealed intriguing ribosome properties challenging this view. Many studies have shown that to perform an accurate protein synthesis in a fluctuating cellular environment, ribosomes sense, transfer information and even make decisions. This complex "behaviour" that goes far beyond the skills of a simple mechanical machine has suggested that the ribosomal protein networks could play a role equivalent to nervous circuits at a molecular scale to enable information transfer and processing during translation. We analyse here the significance of this analogy and establish a preliminary link between two fields: ribosome structure-function studies and the analysis of information processing systems. This cross-disciplinary analysis opens new perspectives about the mechanisms of information transfer and processing in ribosomes and may provide new conceptual frameworks for the understanding of the behaviours of unicellular organisms.
Keywords: complexity; evolution; information processing; nervous circuit; network; neuron; protein interface; ribosome; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



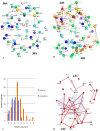


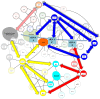
Similar articles
-
Neuron-Like Networks Between Ribosomal Proteins Within the Ribosome.Sci Rep. 2016 May 26;6:26485. doi: 10.1038/srep26485. Sci Rep. 2016. PMID: 27225526 Free PMC article.
-
Towards the Idea of Molecular Brains.Int J Mol Sci. 2021 Nov 1;22(21):11868. doi: 10.3390/ijms222111868. Int J Mol Sci. 2021. PMID: 34769300 Free PMC article. Review.
-
Ribosomal proteomics: Strategies, approaches, and perspectives.Biochimie. 2015 Jun;113:69-77. doi: 10.1016/j.biochi.2015.03.024. Epub 2015 Apr 10. Biochimie. 2015. PMID: 25869001 Review.
-
A structural understanding of the dynamic ribosome machine.Nat Rev Mol Cell Biol. 2008 Mar;9(3):242-53. doi: 10.1038/nrm2352. Nat Rev Mol Cell Biol. 2008. PMID: 18292779 Review.
-
Atomic structures of the eukaryotic ribosome.Trends Biochem Sci. 2012 May;37(5):189-98. doi: 10.1016/j.tibs.2012.02.007. Epub 2012 Mar 20. Trends Biochem Sci. 2012. PMID: 22436288 Review.
Cited by
-
Evolution of ribosomal protein network architectures.Sci Rep. 2021 Jan 12;11(1):625. doi: 10.1038/s41598-020-80194-4. Sci Rep. 2021. PMID: 33436806 Free PMC article.
-
Cellular and Natural Viral Engineering in Cognition-Based Evolution.Commun Integr Biol. 2023 May 2;16(1):2196145. doi: 10.1080/19420889.2023.2196145. eCollection 2023. Commun Integr Biol. 2023. PMID: 37153718 Free PMC article. Review.
-
The Expanding Universe of Extensions and Tails: Ribosomal Proteins and Histones in RNA and DNA Complex Signaling and Dynamics.Genes (Basel). 2025 Jan 1;16(1):45. doi: 10.3390/genes16010045. Genes (Basel). 2025. PMID: 39858592 Free PMC article. Review.
-
Bioluminescence and Photoreception in Unicellular Organisms: Light-Signalling in a Bio-Communication Perspective.Int J Mol Sci. 2021 Oct 20;22(21):11311. doi: 10.3390/ijms222111311. Int J Mol Sci. 2021. PMID: 34768741 Free PMC article. Review.
-
Spatially Enriched Paralog Rearrangements Argue Functionally Diverse Ribosomes Arise during Cold Acclimation in Arabidopsis.Int J Mol Sci. 2021 Jun 7;22(11):6160. doi: 10.3390/ijms22116160. Int J Mol Sci. 2021. PMID: 34200446 Free PMC article.
References
-
- Bullock H., Horridge A. Structure and Function in the Nervous Systems of Invertebrates. W.H. Freeman; San Francisco, CA, USA: 1965.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

