Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging
- PMID: 31207930
- PMCID: PMC6627902
- DOI: 10.3390/cancers11060829
Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging
Abstract
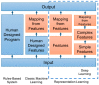
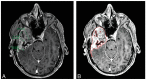
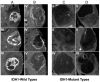
Radiographic assessment with magnetic resonance imaging (MRI) is widely used to characterize gliomas, which represent 80% of all primary malignant brain tumors. Unfortunately, glioma biology is marked by heterogeneous angiogenesis, cellular proliferation, cellular invasion, and apoptosis. This translates into varying degrees of enhancement, edema, and necrosis, making reliable imaging assessment challenging. Deep learning, a subset of machine learning artificial intelligence, has gained traction as a method, which has seen effective employment in solving image-based problems, including those in medical imaging. This review seeks to summarize current deep learning applications used in the field of glioma detection and outcome prediction and will focus on (1) pre- and post-operative tumor segmentation, (2) genetic characterization of tissue, and (3) prognostication. We demonstrate that deep learning methods of segmenting, characterizing, grading, and predicting survival in gliomas are promising opportunities that may enhance both research and clinical activities.
Keywords: artificial intelligence; deep learning; glioblastoma; glioma; machine learning; neural network.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Ostrom Q.T., Gittleman H., Fulop J., Liu M., Blanda R., Kromer C., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl. 4):iv1–iv62. doi: 10.1093/neuonc/nov189. - DOI - PMC - PubMed
-
- Wiki for the VASARI Feature Set the National Cancer Institute Web Site. [(accessed on 10 April 2019)]; Available online: https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Pro....
-
- Gutman D.A., Cooper L.A., Hwang S.N., Holder C.A., Gao J., Aurora T.D., Dunn W.D., Jr., Scarpace L., Mikkelsen T., Jain R., et al. MR imaging predictors of molecular profile and survival: Multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. - DOI - PMC - PubMed

