Euglena Central Metabolic Pathways and Their Subcellular Locations
- PMID: 31207935
- PMCID: PMC6630311
- DOI: 10.3390/metabo9060115
Euglena Central Metabolic Pathways and Their Subcellular Locations
Abstract
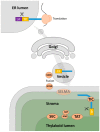
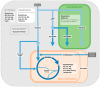
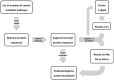
Euglenids are a group of algae of great interest for biotechnology, with a large and complex metabolic capability. To study the metabolic network, it is necessary to know where the component enzymes are in the cell, but despite a long history of research into Euglena, the subcellular locations of many major pathways are only poorly defined. Euglena is phylogenetically distant from other commonly studied algae, they have secondary plastids bounded by three membranes, and they can survive after destruction of their plastids. These unusual features make it difficult to assume that the subcellular organization of the metabolic network will be equivalent to that of other photosynthetic organisms. We analysed bioinformatic, biochemical, and proteomic information from a variety of sources to assess the subcellular location of the enzymes of the central metabolic pathways, and we use these assignments to propose a model of the metabolic network of Euglena. Other than photosynthesis, all major pathways present in the chloroplast are also present elsewhere in the cell. Our model demonstrates how Euglena can synthesise all the metabolites required for growth from simple carbon inputs, and can survive in the absence of chloroplasts.
Keywords: Euglena; central metabolic pathway; subcellular location.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Evolutionary Origin of Euglena.Adv Exp Med Biol. 2017;979:3-17. doi: 10.1007/978-3-319-54910-1_1. Adv Exp Med Biol. 2017. PMID: 28429314 Review.
-
Euglenoid flagellates: a multifaceted biotechnology platform.J Biotechnol. 2015 May 20;202:135-45. doi: 10.1016/j.jbiotec.2014.11.035. Epub 2014 Dec 16. J Biotechnol. 2015. PMID: 25527385 Review.
-
Photo and Nutritional Regulation of Euglena Organelle Development.Adv Exp Med Biol. 2017;979:159-182. doi: 10.1007/978-3-319-54910-1_9. Adv Exp Med Biol. 2017. PMID: 28429322 Review.
-
On the origin of chloroplasts, import mechanisms of chloroplast-targeted proteins, and loss of photosynthetic ability - review.Folia Microbiol (Praha). 2009;54(4):303-21. doi: 10.1007/s12223-009-0048-z. Epub 2009 Oct 14. Folia Microbiol (Praha). 2009. PMID: 19826918 Review.
-
Metabolic quirks and the colourful history of the Euglena gracilis secondary plastid.New Phytol. 2020 Feb;225(4):1578-1592. doi: 10.1111/nph.16237. Epub 2019 Nov 4. New Phytol. 2020. PMID: 31580486
Cited by
-
Glycosylated proteins in the protozoan alga Euglena gracilis: a proteomic approach.FEMS Microbiol Lett. 2023 Jan 17;370:fnac120. doi: 10.1093/femsle/fnac120. FEMS Microbiol Lett. 2023. PMID: 36477252 Free PMC article.
-
Transcriptomic response analysis of ultraviolet mutagenesis combined with high carbon acclimation to promote photosynthetic carbon assimilation in Euglena gracilis.Front Microbiol. 2024 Aug 29;15:1444420. doi: 10.3389/fmicb.2024.1444420. eCollection 2024. Front Microbiol. 2024. PMID: 39268527 Free PMC article.
-
Metabolomic Insights into the Adaptations and Biotechnological Potential of Euglena gracilis Under Different Trophic Conditions.Plants (Basel). 2025 May 22;14(11):1580. doi: 10.3390/plants14111580. Plants (Basel). 2025. PMID: 40508257 Free PMC article.
-
Discovery of a parallel family of euglenatide analogs in Euglena gracilis.Nat Prod Bioprospect. 2025 Jan 6;15(1):10. doi: 10.1007/s13659-024-00490-8. Nat Prod Bioprospect. 2025. PMID: 39760919 Free PMC article.
-
Metabolic network reconstruction of Euglena gracilis: Current state, challenges, and applications.Front Microbiol. 2023 Mar 2;14:1143770. doi: 10.3389/fmicb.2023.1143770. eCollection 2023. Front Microbiol. 2023. PMID: 36937274 Free PMC article. Review.
References
-
- Einicker-Lamas M., Mezian G.A., Fernandes T.B., Silva F.L.S., Guerra F., Miranda K., Attias M., Oliveira M.M. Euglena gracilis as a model for the study of Cu2+ and Zn2+ toxicity and accumulation in eukaryotic cells. Environ. Pollut. 2002;120:779–786. doi: 10.1016/S0269-7491(02)00170-7. - DOI - PubMed
-
- Kitaoka S., Nakano Y., Miyatake K., Yokota A. Enzymes and their functional location. In: Buetow D.E., editor. The Biology of Euglena. Volume 4. Academic Press Limited; London, UK: 1989. pp. 2–119.
-
- Wolken J.J. Euglena: An Experimental Organism for Biochemical and Biophysical Studies. Institute of Microbiology, Rutgers; New Brunswick, NJ, USA: 1961. p. 173.
Grants and funding
LinkOut - more resources
Full Text Sources

