A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates
- PMID: 31208076
- PMCID: PMC6617194
- DOI: 10.3390/jcm8060858
A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates
Abstract
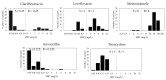
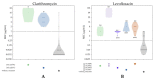
We evaluated the primary resistance of Helicobacter pylori (H. pylori) to routinely used antibiotics in Cambodia, an unexplored topic in the country, and assessed next-generation sequencing's (NGS) potential to discover genetic resistance determinants. Fifty-five H. pylori strains were successfully cultured and screened for antibiotic susceptibility using agar dilution. Genotypic analysis was performed using NGS data with a CLC genomic workbench. PlasmidSeeker was used to detect plasmids. The correlation between resistant genotypes and phenotypes was evaluated statistically. Resistances to metronidazole (MTZ), levofloxacin (LVX), clarithromycin (CLR), and amoxicillin (AMX) were 96.4%, 67.3%, 25.5%, and 9.1%, respectively. No resistance to tetracycline (TET) was observed. Multi-drug resistance affected 76.4% of strains. No plasmids were found, but genetic determinants of resistance to CLR, LVX, and AMX were 23S rRNA (A2146G and A2147G), GyrA (N87K and D91Y/N/G), and pbp1 (P473L), respectively. No determinants were genetically linked to MTZ or TET resistance. There was high concordance between resistant genotypes and phenotypes for AMX, LVX, and CLR. We observed high antibiotic resistance rates of CLR, MTZ, and LVX, emphasizing the need for periodic evaluation and alternative therapies in Cambodia. NGS showed high capability for detecting genetic resistance determinants and potential for implementation in local treatment policies.
Keywords: Helicobacter pylori; antibiotic resistance; genetic determinants; mutation; next-generation sequencing; whole genome sequence.
Conflict of interest statement
All authors declare that they have no potential conflict of interest.
Figures

References
-
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

