Analysis of allele-specific expression using RNA-seq of the Korean native pig and Landrace reciprocal cross
- PMID: 31208168
- PMCID: PMC6819674
- DOI: 10.5713/ajas.19.0097
Analysis of allele-specific expression using RNA-seq of the Korean native pig and Landrace reciprocal cross
Abstract
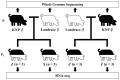
Objective: We tried to analyze allele-specific expression in the pig neocortex using bioinformatic analysis of high-throughput sequencing results from the parental genomes and offspring transcriptomes from reciprocal crosses between Korean Native and Landrace pigs.
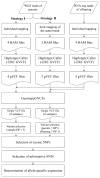
Methods: We carried out sequencing of parental genomes and offspring transcriptomes using next generation sequencing. We subsequently carried out genome scale identification of SNPs in two different ways using either individual genome mapping or joint genome mapping of the same breed parents that were used for the reciprocal crosses. Using parent-specific SNPs, allele-specifically expressed genes were analyzed.
Results: Because of the low genome coverage (~4x) of the sequencing results, most SNPs were non-informative for parental lineage determination of the expressed alleles in the offspring and were thus excluded from our analysis. Consequently, 436 SNPs covering 336 genes were applicable to measure the imbalanced expression of paternal alleles in the offspring. By calculating the read ratios of parental alleles in the offspring, we identified seven genes showing allele-biased expression (P < 0.05) including three previously reported and four newly identified genes in this study.
Conclusion: The newly identified allele-specifically expressing genes in the neocortex of pigs should contribute to improving our knowledge on genomic imprinting in pigs. To our knowledge, this is the first study of allelic imbalance using high throughput analysis of both parental genomes and offspring transcriptomes of the reciprocal cross in outbred animals. Our study also showed the effect of the number of informative animals on the genome level investigation of allele-specific expression using RNA-seq analysis in livestock species.
Keywords: Allele-specific Expression (ASE); Genomic Imprinting; Korean Native Pigs; Reciprocal Cross; Swine.
Conflict of interest statement
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Figures

Similar articles
-
Genome-wide identification of imprinted genes in pigs and their different imprinting status compared with other mammals.Zool Res. 2020 Nov 18;41(6):721-725. doi: 10.24272/j.issn.2095-8137.2020.072. Zool Res. 2020. PMID: 32808516 Free PMC article.
-
Extensive variation between tissues in allele specific expression in an outbred mammal.BMC Genomics. 2015 Nov 23;16:993. doi: 10.1186/s12864-015-2174-0. BMC Genomics. 2015. PMID: 26596891 Free PMC article.
-
Variant calling from RNA-seq data of the brain transcriptome of pigs and its application for allele-specific expression and imprinting analysis.Gene. 2018 Jan 30;641:367-375. doi: 10.1016/j.gene.2017.10.076. Epub 2017 Oct 27. Gene. 2018. PMID: 29111207
-
Using next-generation RNA sequencing to identify imprinted genes.Heredity (Edinb). 2014 Aug;113(2):156-66. doi: 10.1038/hdy.2014.18. Epub 2014 Mar 12. Heredity (Edinb). 2014. PMID: 24619182 Free PMC article. Review.
-
Parent-of-origin specific QTL--a possibility towards understanding reciprocal effects in chicken and the origin of imprinting.Cytogenet Genome Res. 2007;117(1-4):305-12. doi: 10.1159/000103192. Cytogenet Genome Res. 2007. PMID: 17675872 Review.
Cited by
-
Multi-omic characterization of allele-specific regulatory variation in hybrid pigs.Nat Commun. 2024 Jul 3;15(1):5587. doi: 10.1038/s41467-024-49923-5. Nat Commun. 2024. PMID: 38961076 Free PMC article.
-
Genome-wide identification of imprinted genes in pigs and their different imprinting status compared with other mammals.Zool Res. 2020 Nov 18;41(6):721-725. doi: 10.24272/j.issn.2095-8137.2020.072. Zool Res. 2020. PMID: 32808516 Free PMC article.
-
Screening of Genes Related to Fat Deposition of Pekin Ducks Based on Transcriptome Analysis.Animals (Basel). 2024 Jan 15;14(2):268. doi: 10.3390/ani14020268. Animals (Basel). 2024. PMID: 38254437 Free PMC article.
-
Effects of the FHL2 gene on the development of subcutaneous and intramuscular adipocytes in goats.BMC Genomics. 2024 Sep 11;25(1):850. doi: 10.1186/s12864-024-10755-8. BMC Genomics. 2024. PMID: 39261767 Free PMC article.
References
LinkOut - more resources
Full Text Sources

