GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction
- PMID: 31208283
- PMCID: PMC7138202
- DOI: 10.1080/15548627.2019.1632620
GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction
Abstract
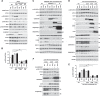
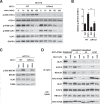
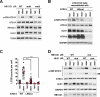
ULK1 (unc-51 like autophagy activating kinase 1) is the key mediator of MTORC1 signaling to macroautophagy/autophagy. ULK1 functions as a protein complex by interacting with ATG13, RB1CC1/FIP200, and ATG101. How the ULK1 complex is regulated to trigger autophagy induction remains unclear. In this study, we have determined roles of Atg8-family proteins (ATG8s) in regulating ULK1 activity and autophagy. Using human cells depleted of each subfamily of ATG8, we found that the GABARAP subfamily positively regulates ULK1 activity and phagophore and autophagosome formation in response to starvation. In contrast, the LC3 subfamily negatively regulates ULK1 activity and phagophore formation. By reconstituting ATG8-depleted cells with individual ATG8 members, we identified GABARAP and GABARAPL1 as positive and LC3B and LC3C as negative regulators of ULK1 activity. To address the role of ATG8 binding to ULK1, we mutated the LIR of endogenous ULK1 to disrupt the ATG8-ULK1 interaction by genome editing. The mutation drastically reduced the activity of ULK1, autophagic degradation of SQSTM1, and phagophore formation in response to starvation. The mutation also suppressed the formation and turnover of autophagosomes in response to starvation. Similar to the mutation of the ULK1 LIR, disruption of the ATG13-ATG8 interaction suppressed ULK1 activity and autophagosome formation. In contrast, RB1CC1 did not show any specific binding to ATG8s, and mutation of its LIR did not affect ULK1 activity. Together, this study demonstrates differential binding and opposite regulation of the ULK1 complex by GABARAPs and LC3s, and an important role of the ULK1- and ATG13-ATG8 interactions in autophagy induction.Abbreviations: ATG5: autophagy related 5; ATG7: autophagy related 7; ATG8: autophagy related 8; ATG13: autophagy related 13; ATG14: autophagy related 14; ATG16L1: autophagy related 16 like 1; ATG101: autophagy related 101; BAFA1: bafilomycin A1; BECN1: beclin 1; Cas9: CRISPR associated protein 9; CRISPR: clustered regularly interspaced short palindromic repeats; EBSS: earle's balanced salt solution; DAPI: 4'-6-diamidino-2-phenylindole; GABARAP: GABA type A receptor-associated protein; GABARAPL1: GABA type A receptor-associated protein like 1; GABARAPL2: GABA type A receptor-associated protein like 2; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GFP: green fluorescence protein; gRNA: guide RNA; KI: kinase inactive mutant; KO: knockout; LC3A: microtubule associated protein 1 light chain 3 alpha; LC3B: microtubule associated protein 1 light chain 3 beta; LC3C: microtubule associated protein 1 light chain 3 gamma; LIR: LC3-interacting region; MTORC1: mechanistic target of rapamycin kinase complex 1; PBS: phosphate buffered saline; PCR: polymerase chain reaction; PE: phosphatidylethanolamine; PtdIns3P: phosphatidylinositol-3-phosphate; qPCR: quantitative PCR; RB1CC1/FIP200: RB1 inducible coiled-coil 1; RPS6KB1: ribosomal protein S6 kinase B1; SEM: standard error of the mean; SQSTM1/p62: sequestosome 1; TALEN: transcription activator-like effector nuclease; TUBA: tubulin alpha; ULK1: unc-51 like autophagy activating kinase 1; WB: western blotting; WIPI2: WD repeat domain phosphoinositide interacting 2; WT: wild type.
Keywords: ATG8; GABARAP; LC3; LIR; ULK1.
Figures




Similar articles
-
Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.Autophagy. 2019 Aug;15(8):1333-1355. doi: 10.1080/15548627.2019.1581009. Epub 2019 Mar 4. Autophagy. 2019. PMID: 30767700 Free PMC article.
-
Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells.Autophagy. 2019 Jun;15(6):976-997. doi: 10.1080/15548627.2019.1569925. Epub 2019 Feb 1. Autophagy. 2019. PMID: 30661429 Free PMC article.
-
Artificial targeting of autophagy components to mitochondria reveals both conventional and unconventional mitophagy pathways.Autophagy. 2025 Feb;21(2):315-337. doi: 10.1080/15548627.2024.2395149. Epub 2024 Sep 8. Autophagy. 2025. PMID: 39177530 Free PMC article.
-
Autophagy in the physiological endometrium and cancer.Autophagy. 2021 May;17(5):1077-1095. doi: 10.1080/15548627.2020.1752548. Epub 2020 May 13. Autophagy. 2021. PMID: 32401642 Free PMC article. Review.
-
The functions of Atg8-family proteins in autophagy and cancer: linked or unrelated?Autophagy. 2021 Mar;17(3):599-611. doi: 10.1080/15548627.2020.1749367. Epub 2020 Apr 19. Autophagy. 2021. PMID: 32255730 Free PMC article. Review.
Cited by
-
The DNA methyltransferase DNMT3A contributes to autophagy long-term memory.Autophagy. 2021 May;17(5):1259-1277. doi: 10.1080/15548627.2020.1816664. Epub 2020 Sep 14. Autophagy. 2021. PMID: 32876528 Free PMC article.
-
Exosomal circHIPK3 derived from umbilical cord-derived mesenchymal stem cells enhances skin fibroblast autophagy by blocking miR-20b-5p/ULK1/Atg13 axis.J Diabetes Investig. 2023 Dec;14(12):1344-1355. doi: 10.1111/jdi.14077. Epub 2023 Sep 8. J Diabetes Investig. 2023. PMID: 37688345 Free PMC article.
-
Stapled Peptide Inhibitors of Autophagy Adapter LC3B.Chembiochem. 2020 Oct 1;21(19):2777-2785. doi: 10.1002/cbic.202000212. Epub 2020 Jun 22. Chembiochem. 2020. PMID: 32406996 Free PMC article.
-
Autophagosome biogenesis and human health.Cell Discov. 2020 Jun 2;6(1):33. doi: 10.1038/s41421-020-0166-y. eCollection 2020. Cell Discov. 2020. PMID: 32528724 Free PMC article. Review.
-
Disulfidptosis: A new type of cell death.Apoptosis. 2024 Oct;29(9-10):1309-1329. doi: 10.1007/s10495-024-01989-8. Epub 2024 Jun 17. Apoptosis. 2024. PMID: 38886311 Free PMC article. Review.
References
-
- Mercer CA, Kaliappan A, Dennis PB.. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009. July;5(5):649–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
