A simplified transposon mutagenesis method to perform phenotypic forward genetic screens in cultured cells
- PMID: 31208320
- PMCID: PMC6580595
- DOI: 10.1186/s12864-019-5888-6
A simplified transposon mutagenesis method to perform phenotypic forward genetic screens in cultured cells
Abstract
Background: The introduction of genome-wide shRNA and CRISPR libraries has facilitated cell-based screens to identify loss-of-function mutations associated with a phenotype of interest. Approaches to perform analogous gain-of-function screens are less common, although some reports have utilized arrayed viral expression libraries or the CRISPR activation system. However, a variety of technical and logistical challenges make these approaches difficult for many labs to execute. In addition, genome-wide shRNA or CRISPR libraries typically contain of hundreds of thousands of individual engineered elements, and the associated complexity creates issues with replication and reproducibility for these methods.
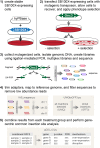
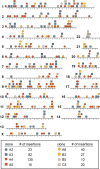
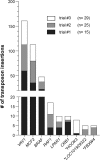
Results: Here we describe a simple, reproducible approach using the SB transposon system to perform phenotypic cell-based genetic screens. This approach employs only three plasmids to perform unbiased, whole-genome transposon mutagenesis. We also describe a ligation-mediated PCR method that can be used in conjunction with the included software tools to map raw sequence data, identify candidate genes associated with phenotypes of interest, and predict the impact of recurrent transposon insertions on candidate gene function. Finally, we demonstrate the high reproducibility of our approach by having three individuals perform independent replicates of a mutagenesis screen to identify drivers of vemurafenib resistance in cultured melanoma cells.
Conclusions: Collectively, our work establishes a facile, adaptable method that can be performed by labs of any size to perform robust, genome-wide screens to identify genes that influence phenotypes of interest.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Identification of Sleeping Beauty transposon insertions in solid tumors using linker-mediated PCR.J Vis Exp. 2013 Feb 1;(72):e50156. doi: 10.3791/50156. J Vis Exp. 2013. PMID: 23407503 Free PMC article.
-
Ex Vivo Transposon-Mediated Genetic Screens for Cancer Gene Discovery.Methods Mol Biol. 2019;1907:145-157. doi: 10.1007/978-1-4939-8967-6_12. Methods Mol Biol. 2019. PMID: 30542998 Free PMC article.
-
Transposon-mediated mutagenesis of somatic cells in the mouse for cancer gene identification.Methods. 2009 Nov;49(3):282-6. doi: 10.1016/j.ymeth.2009.07.002. Epub 2009 Jul 14. Methods. 2009. PMID: 19607923 Free PMC article.
-
Insertional engineering of chromosomes with Sleeping Beauty transposition: an overview.Methods Mol Biol. 2011;738:69-85. doi: 10.1007/978-1-61779-099-7_5. Methods Mol Biol. 2011. PMID: 21431720 Review.
-
Validation-Based Insertional Mutagenesis (VBIM), A Powerful Forward Genetic Screening Strategy.Curr Protoc. 2022 Mar;2(3):e394. doi: 10.1002/cpz1.394. Curr Protoc. 2022. PMID: 35316583 Free PMC article. Review.
Cited by
-
Transmicron: accurate prediction of insertion probabilities improves detection of cancer driver genes from transposon mutagenesis screens.Nucleic Acids Res. 2023 Feb 28;51(4):e21. doi: 10.1093/nar/gkac1215. Nucleic Acids Res. 2023. PMID: 36617985 Free PMC article.
-
Understanding cancer drug resistance with Sleeping Beauty functional genomic screens: Application to MAPK inhibition in cutaneous melanoma.iScience. 2023 Aug 31;26(10):107805. doi: 10.1016/j.isci.2023.107805. eCollection 2023 Oct 20. iScience. 2023. PMID: 37860756 Free PMC article.
-
Single-cell genomics analysis reveals complex genetic interactions in an in vivo model of acquired BRAF inhibitor resistance.NAR Cancer. 2024 Jan 11;6(1):zcad061. doi: 10.1093/narcan/zcad061. eCollection 2024 Mar. NAR Cancer. 2024. PMID: 38213996 Free PMC article.
-
A Genetic Screen to Identify Gain- and Loss-of-Function Modifications that Enhance T-cell Infiltration into Tumors.Cancer Immunol Res. 2020 Sep;8(9):1206-1214. doi: 10.1158/2326-6066.CIR-20-0056. Epub 2020 Jul 1. Cancer Immunol Res. 2020. PMID: 32611665 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

