Machine learning with the TCGA-HNSC dataset: improving usability by addressing inconsistency, sparsity, and high-dimensionality
- PMID: 31208324
- PMCID: PMC6580485
- DOI: 10.1186/s12859-019-2929-8
Machine learning with the TCGA-HNSC dataset: improving usability by addressing inconsistency, sparsity, and high-dimensionality
Abstract
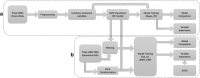
Background: In the era of precision oncology and publicly available datasets, the amount of information available for each patient case has dramatically increased. From clinical variables and PET-CT radiomics measures to DNA-variant and RNA expression profiles, such a wide variety of data presents a multitude of challenges. Large clinical datasets are subject to sparsely and/or inconsistently populated fields. Corresponding sequencing profiles can suffer from the problem of high-dimensionality, where making useful inferences can be difficult without correspondingly large numbers of instances. In this paper we report a novel deployment of machine learning techniques to handle data sparsity and high dimensionality, while evaluating potential biomarkers in the form of unsupervised transformations of RNA data. We apply preprocessing, MICE imputation, and sparse principal component analysis (SPCA) to improve the usability of more than 500 patient cases from the TCGA-HNSC dataset for enhancing future oncological decision support for Head and Neck Squamous Cell Carcinoma (HNSCC).
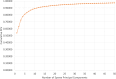
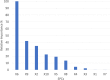
Results: Imputation was shown to improve prognostic ability of sparse clinical treatment variables. SPCA transformation of RNA expression variables reduced runtime for RNA-based models, though changes to classifier performance were not significant. Gene ontology enrichment analysis of gene sets associated with individual sparse principal components (SPCs) are also reported, showing that both high- and low-importance SPCs were associated with cell death pathways, though the high-importance gene sets were found to be associated with a wider variety of cancer-related biological processes.
Conclusions: MICE imputation allowed us to impute missing values for clinically informative features, improving their overall importance for predicting two-year recurrence-free survival by incorporating variance from other clinical variables. Dimensionality reduction of RNA expression profiles via SPCA reduced both computation cost and model training/evaluation time without affecting classifier performance, allowing researchers to obtain experimental results much more quickly. SPCA simultaneously provided a convenient avenue for consideration of biological context via gene ontology enrichment analysis.
Keywords: Decision support; Dimensionality reduction; Gene ontology enrichment analysis; Machine learning; Unsupervised transformation; hnscc; tcga.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Liu J, Wu Y, Wang Q, Liu X, Liao X, Pan J. Bioinformatic analysis of PFN2 dysregulation and its prognostic value in head and neck squamous carcinoma. (1744–8301 (Electronic)). 2018. - PubMed
-
- Huang H, Lin C, Yang C, Ho C, Chang Y, Chang J, editors. An integrative analysis for Cancer studies. 2016 IEEE 16th international conference on bioinformatics and bioengineering (BIBE); 2016 31 Oct.-2 Nov. 2016.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

