Combining learning and constraints for genome-wide protein annotation
- PMID: 31208327
- PMCID: PMC6580517
- DOI: 10.1186/s12859-019-2875-5
Combining learning and constraints for genome-wide protein annotation
Abstract
Background: The advent of high-throughput experimental techniques paved the way to genome-wide computational analysis and predictive annotation studies. When considering the joint annotation of a large set of related entities, like all proteins of a certain genome, many candidate annotations could be inconsistent, or very unlikely, given the existing knowledge. A sound predictive framework capable of accounting for this type of constraints in making predictions could substantially contribute to the quality of machine-generated annotations at a genomic scale.
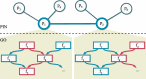
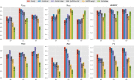
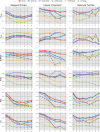
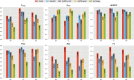
Results: We present OCELOT, a predictive pipeline which simultaneously addresses functional and interaction annotation of all proteins of a given genome. The system combines sequence-based predictors for functional and protein-protein interaction (PPI) prediction with a consistency layer enforcing (soft) constraints as fuzzy logic rules. The enforced rules represent the available prior knowledge about the classification task, including taxonomic constraints over each GO hierarchy (e.g. a protein labeled with a GO term should also be labeled with all ancestor terms) as well as rules combining interaction and function prediction. An extensive experimental evaluation on the Yeast genome shows that the integration of prior knowledge via rules substantially improves the quality of the predictions. The system largely outperforms GoFDR, the only high-ranking system at the last CAFA challenge with a readily available implementation, when GoFDR is given access to intra-genome information only (as OCELOT), and has comparable or better results (depending on the hierarchy and performance measure) when GoFDR is allowed to use information from other genomes. Our system also compares favorably to recent methods based on deep learning.
Keywords: Genome annotation; Kernel methods; Protein function prediction; Protein-protein interaction.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Term Matrix: a novel Gene Ontology annotation quality control system based on ontology term co-annotation patterns.Open Biol. 2020 Sep;10(9):200149. doi: 10.1098/rsob.200149. Epub 2020 Sep 2. Open Biol. 2020. PMID: 32875947 Free PMC article.
-
False positive reduction in protein-protein interaction predictions using gene ontology annotations.BMC Bioinformatics. 2007 Jul 23;8:262. doi: 10.1186/1471-2105-8-262. BMC Bioinformatics. 2007. PMID: 17645798 Free PMC article.
-
Saccharomyces cerevisiae: gene annotation and genome variability, state of the art through comparative genomics.Methods Mol Biol. 2011;759:31-40. doi: 10.1007/978-1-61779-173-4_2. Methods Mol Biol. 2011. PMID: 21863479
-
How to overcome constraints imposed by microsporidian genome features to ensure gene prediction?J Eukaryot Microbiol. 2024 Sep-Oct;71(5):e13038. doi: 10.1111/jeu.13038. Epub 2024 Jun 27. J Eukaryot Microbiol. 2024. PMID: 38934348 Review.
-
Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review.
References
-
- Friedberg I. Automated protein function prediction–the genomic challenge. Brief Bioinform. 2006;7(3):225–42. - PubMed
-
- Keskin O, Gursoy A, Ma B, Nussinov R, et al. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev. 2008;108(4):1225–44. - PubMed
-
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–90. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

