Core encoding sequences of Hepatitis C virus in Ghanaian blood donors are predominantly mosaics of different genotype 2 strains and cannot distinguish subtypes
- PMID: 31208352
- PMCID: PMC6580569
- DOI: 10.1186/s12879-019-4155-4
Core encoding sequences of Hepatitis C virus in Ghanaian blood donors are predominantly mosaics of different genotype 2 strains and cannot distinguish subtypes
Abstract
Background: Distribution of Hepatitis C virus (HCV) genotypes varies significantly worldwide. Genomic diversity between genotypes has implications for treatment, vaccine development and optimal design of HCV diagnostic assays. Molecular characterization of HCV in different geographical areas is therefore very essential for management and public health control of HCV infection. This study investigated the molecular epidemiology and characteristics of HCV genotypes in healthy individuals in Accra, Ghana.
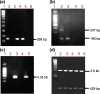
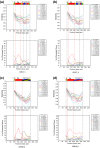
Methods: An experimental study was carried out on blood samples obtained from voluntary blood donors. Two hundred samples were initially screened for HCV antibodies and infection was confirmed by RNA detection through RT-PCR of the 5'-untranslated region (5'UTR). The core gene sequences were analysed for HCV genotype determination by genotype-specific PCR; and then by cloning and direct sequencing followed by phylogenetic analysis. The sequences were further analysed in detail by similarity plotting.
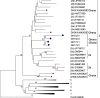
Results: Molecular diagnosis confirmed the presence of HCV RNA in 2 out of 200 (1%) blood donors. Initial genotyping by genotype-specific PCR identified all two infections as subtypes 2a and 2b of genotype 2. Extensive evolutionary and genetic analyses indicated two epidemiological profiles. First, phylogenetic tree topologies clearly showed that, collectively, the core sequences of the Ghanaian HCV isolates belong to a single, distinct genetic group within HCV genotype 2 cluster, with high genetic similarity and rapid sequence variation in a single individual. Second, the sequences are mosaics comprising 2e and other genotype 2 subtype fragments. The analyses underscore a unique and complex HCV genotype 2 core sequence profile of the Ghanaian isolates.
Conclusions: Analysis of HCV core encoding sequences from Ghanaian blood donors in Accra confirmed predominance of genotype 2 HCV among healthy individuals. However, the isolates could not be classified into subtypes, possibly due to their complex sequence pattern that might suggest high mutability of the prevailing genotype. The core region of Ghanaian HCV therefore may not be suitable for distinguishing subtypes. These findings extend those from previous studies and thus underscore the need to search for subtype-informative region of Ghanaian HCV to elucidate the genetic diversity and factors determining outcome of HCV infections in Ghana.
Keywords: Blood donors; Core gene; Genotype; Ghana; HCV; Molecular epidemiology; Seroprevalence.
Conflict of interest statement
The authors declare that there are no competing interests.
Figures
Similar articles
-
Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa.J Virol. 2003 Jul;77(14):7914-23. doi: 10.1128/jvi.77.14.7914-7923.2003. J Virol. 2003. PMID: 12829831 Free PMC article.
-
Molecular characterization of hepatitis C virus genotype 6 subtypes in Thai blood donors.J Microbiol Immunol Infect. 2017 Feb;50(1):26-31. doi: 10.1016/j.jmii.2015.01.006. Epub 2015 Jan 30. J Microbiol Immunol Infect. 2017. PMID: 25731701
-
Molecular characterization, distribution, and dynamics of hepatitis C virus genotypes in blood donors in Colombia.J Med Virol. 2010 Nov;82(11):1889-98. doi: 10.1002/jmv.21908. J Med Virol. 2010. PMID: 20872715
-
[The different epidemic and evolution of HCV genotypes].Yi Chuan. 2012 Jun;34(6):666-72. doi: 10.3724/sp.j.1005.2012.00666. Yi Chuan. 2012. PMID: 22698736 Review. Chinese.
-
Review: molecular epidemiology of hepatitis C virus.J Gastroenterol Hepatol. 1997 Jul;12(7):522-7. doi: 10.1111/j.1440-1746.1997.tb00477.x. J Gastroenterol Hepatol. 1997. PMID: 9257244 Review.
Cited by
-
'Unusual' HCV genotype subtypes: origin, distribution, sensitivity to direct-acting antiviral drugs and behaviour on antiviral treatment and retreatment.Gut. 2024 Aug 8;73(9):1570-1582. doi: 10.1136/gutjnl-2024-332177. Gut. 2024. PMID: 38782565 Free PMC article. Review.
References
-
- World Health Organization. Know hepatitis - act now. 2/9/2018. http://www.who.int/hepatitis/en/. Accessed 9 Feb 2018.
-
- WHO. Hepatitis C. https://www.who.int/en/news-room/fact-sheets/detail/hepatitis-c.
-
- Kurstak E, Kurstak C, Hossain A. The molecular genetics of Hepatitis C virus (HCV) and its use in the diagnosis of HCV infections. In: Kurstak E, editor. New diagnostic procedures. New York: Plenum Medical Book Co; 1994. pp. 59–71.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

