Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue
- PMID: 31208411
- PMCID: PMC6580566
- DOI: 10.1186/s12920-019-0538-z
Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue
Abstract
Background: RNA-seq is poised to play a major role in the management of kidney transplant patients. Rigorous definition of housekeeping genes (HKG) is essential for further progress in this field. Using single genes or a limited set HKG is inherently problematic since their expression might be altered by specific diseases in the patients being studied.
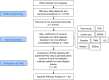
Methods: To generate a HKG set specific for kidney transplantation, we performed RNA-sequencing from renal allograft biopsies collected in a variety of clinical settings. Various normalization methods were applied to identify transcripts that had a coefficient of variation of expression that was below the 2nd percentile across all samples, and the corresponding genes were designated as housekeeping genes. Comparison with transcriptomic data from the Gene Expression Omnibus (GEO) database, pathway analysis and molecular biological functions were utilized to validate the housekeeping genes set.
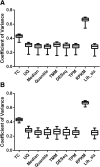
Results: We have developed a bioinformatics solution to this problem by using nine different normalization methods to derive large HKG gene sets from a RNA-seq data set of 47,611 transcripts derived from 30 biopsies. These biopsies were collected in a variety of clinical settings, including normal function, acute rejection, interstitial nephritis, interstitial fibrosis/tubular atrophy and polyomavirus nephropathy. Transcripts with coefficient of variation below the 2nd percentile were designated as HKG, and validated by showing their virtual absence in diseased allograft derived transcriptomic data sets available in the GEO. Pathway analysis indicated a role for these genes in maintenance of cell morphology, pyrimidine metabolism, and intracellular protein signaling.
Conclusions: Utilization of these objectively defined HKG data sets will guard against errors resulting from focusing on individual genes like 18S RNA, actin & tubulin, which do not maintain constant expression across the known spectrum of renal allograft pathology.
Keywords: Genes with housekeeping functions; Kidney transplantation; RNA-sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Deciphering the sex bias in housekeeping gene expression in adipose tissue: a comprehensive meta-analysis of transcriptomic studies.Biol Sex Differ. 2023 Apr 18;14(1):20. doi: 10.1186/s13293-023-00506-x. Biol Sex Differ. 2023. PMID: 37072826 Free PMC article.
-
Urinary cell transcriptomics and acute rejection in human kidney allografts.JCI Insight. 2020 Feb 27;5(4):e131552. doi: 10.1172/jci.insight.131552. JCI Insight. 2020. PMID: 32102984 Free PMC article.
-
Validation of Suitable Housekeeping Genes for the Normalization of mRNA Expression for Studying Tumor Acidosis.Int J Mol Sci. 2018 Sep 26;19(10):2930. doi: 10.3390/ijms19102930. Int J Mol Sci. 2018. PMID: 30261649 Free PMC article.
-
Urinary Cell mRNA Profiles Predictive of Human Kidney Allograft Status.Clin J Am Soc Nephrol. 2021 Oct;16(10):1565-1577. doi: 10.2215/CJN.14010820. Epub 2021 Apr 27. Clin J Am Soc Nephrol. 2021. PMID: 33906907 Free PMC article. Review.
-
Diagnosis of interstitial fibrosis and tubular atrophy in kidney allograft: implementation of microRNAs.Iran J Kidney Dis. 2014 Jan;8(1):4-12. Iran J Kidney Dis. 2014. PMID: 24413714 Review.
Cited by
-
Age-invariant genes: multi-tissue identification and characterization of murine reference genes.Aging (Albany NY). 2025 Jan 27;17(1):170-202. doi: 10.18632/aging.206192. Epub 2025 Jan 27. Aging (Albany NY). 2025. PMID: 39873648 Free PMC article.
-
SAREV: A review on statistical analytics of single-cell RNA sequencing data.Wiley Interdiscip Rev Comput Stat. 2022 Jul-Aug;14(4):e1558. doi: 10.1002/wics.1558. Epub 2021 May 20. Wiley Interdiscip Rev Comput Stat. 2022. PMID: 36034329 Free PMC article.
-
RNA sequencing of whole blood in dogs with primary immune-mediated hemolytic anemia (IMHA) reveals novel insights into disease pathogenesis.PLoS One. 2020 Oct 22;15(10):e0240975. doi: 10.1371/journal.pone.0240975. eCollection 2020. PLoS One. 2020. PMID: 33091028 Free PMC article.
-
HIF regulates multiple translated endogenous retroviruses: Implications for cancer immunotherapy.Cell. 2025 Apr 3;188(7):1807-1827.e34. doi: 10.1016/j.cell.2025.01.046. Epub 2025 Feb 28. Cell. 2025. PMID: 40023154 Free PMC article.
-
Age-Invariant Genes: Multi-Tissue Identification and Characterization of Murine Reference Genes.bioRxiv [Preprint]. 2024 Apr 13:2024.04.09.588721. doi: 10.1101/2024.04.09.588721. bioRxiv. 2024. Update in: Aging (Albany NY). 2025 Jan 27;17(1):170-202. doi: 10.18632/aging.206192. PMID: 38645168 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

