Genomewide Elucidation of Drug Resistance Mechanisms for Systemically Used Antifungal Drugs Amphotericin B, Caspofungin, and Voriconazole in the Budding Yeast
- PMID: 31209012
- PMCID: PMC6709451
- DOI: 10.1128/AAC.02268-18
Genomewide Elucidation of Drug Resistance Mechanisms for Systemically Used Antifungal Drugs Amphotericin B, Caspofungin, and Voriconazole in the Budding Yeast
Abstract
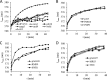
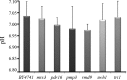
There are only a few antifungal drugs used systemically in treatment, and invasive fungal infections that are resistant to these drugs are an emerging problem in health care. In this study, we performed a high-copy-number genomic DNA (gDNA) library screening to find and characterize genes that reduce susceptibility to amphotericin B, caspofungin, and voriconazole in Saccharomyces cerevisiae We identified the PDR16 and PMP3 genes for amphotericin B, the RMD9 and SWH1 genes for caspofungin, and the MRS3 and TRI1 genes for voriconazole. The deletion mutants for PDR16 and PMP3 were drug susceptible, but the other mutants had no apparent susceptibility. Quantitative-PCR analyses suggested that the corresponding drugs upregulated expression of the PDR16, PMP3, SWH1, and MRS3 genes. To further characterize these genes, we also profiled the global expression patterns of the cells after treatment with the antifungals and determined the genes and paths that were up- or downregulated. We also cloned Candida albicans homologs of the PDR16, PMP3, MRS3, and TRI1 genes and expressed them in S. cerevisiae Heterologous expression of Candida homologs also provided reduced drug susceptibility to the budding yeast cells. Our analyses suggest the involvement of new genes in antifungal drug resistance.
Keywords: amphotericin B; antifungal agents; caspofungin; drug resistance; genomics; multidrug resistance; voriconazole.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- George MJ, Snydman DR, Werner BG, Griffith J, Falagas ME, Dougherty NN, Rubin RH, Boston Center for Liver Transplantation CMVIG-Study Group, Cytogam, MedImmune, Inc. Gaithersburg, Maryland. 1997. The independent role of cytomegalovirus as a risk factor for invasive fungal disease in orthotopic liver transplant recipients. Am J Med 103:106–113. doi:10.1016/S0002-9343(97)80021-6. - DOI - PubMed
-
- De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy, Infectious Diseases Mycoses Study Group Consensus Group. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi:10.1086/588660. - DOI - PMC - PubMed
-
- Lewis RE, Cahyame-Zuniga L, Leventakos K, Chamilos G, Ben-Ami R, Tamboli P, Tarrand J, Bodey GP, Luna M, Kontoyiannis DP. 2013. Epidemiology and sites of involvement of invasive fungal infections in patients with haematological malignancies: a 20-year autopsy study. Mycoses 56:638–645. doi:10.1111/myc.12081. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

