Molecular form and function of the cytokinetic ring
- PMID: 31209062
- PMCID: PMC6602304
- DOI: 10.1242/jcs.226928
Molecular form and function of the cytokinetic ring
Abstract
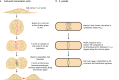
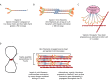
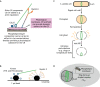
Animal cells, amoebas and yeast divide using a force-generating, actin- and myosin-based contractile ring or 'cytokinetic ring' (CR). Despite intensive research, questions remain about the spatial organization of CR components, the mechanism by which the CR generates force, and how other cellular processes are coordinated with the CR for successful membrane ingression and ultimate cell separation. This Review highlights new findings about the spatial relationship of the CR to the plasma membrane and the arrangement of molecules within the CR from studies using advanced microscopy techniques, as well as mechanistic information obtained from in vitro approaches. We also consider advances in understanding coordinated cellular processes that impact the architecture and function of the CR.
Keywords: Actomyosin ring; Amoeba; Animal cells; Cell division; Contractile ring; Cytokinesis; Cytokinetic ring; Fungi; Metazoa; Yeast.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

