C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa
- PMID: 31209071
- PMCID: PMC6683998
- DOI: 10.1084/jem.20190009
C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa
Abstract
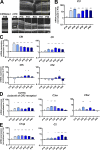
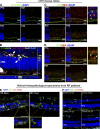
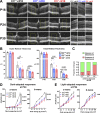
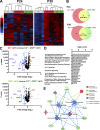
Complement activation has been implicated as contributing to neurodegeneration in retinal and brain pathologies, but its role in retinitis pigmentosa (RP), an inherited and largely incurable photoreceptor degenerative disease, is unclear. We found that multiple complement components were markedly up-regulated in retinas with human RP and the rd10 mouse model, coinciding spatiotemporally with photoreceptor degeneration, with increased C3 expression and activation localizing to activated retinal microglia. Genetic ablation of C3 accelerated structural and functional photoreceptor degeneration and altered retinal inflammatory gene expression. These phenotypes were recapitulated by genetic deletion of CR3, a microglia-expressed receptor for the C3 activation product iC3b, implicating C3-CR3 signaling as a regulator of microglia-photoreceptor interactions. Deficiency of C3 or CR3 decreased microglial phagocytosis of apoptotic photoreceptors and increased microglial neurotoxicity to photoreceptors, demonstrating a novel adaptive role for complement-mediated microglial clearance of apoptotic photoreceptors in RP. These homeostatic neuroinflammatory mechanisms are relevant to the design and interpretation of immunomodulatory therapeutic approaches to retinal degenerative disease.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures



References
-
- Bosco A., Anderson S.R., Breen K.T., Romero C.O., Steele M.R., Chiodo V.A., Boye S.L., Hauswirth W.W., Tomlinson S., and Vetter M.L.. 2018. Complement C3-Targeted Gene Therapy Restricts Onset and Progression of Neurodegeneration in Chronic Mouse Glaucoma. Mol. Ther. 26:2379–2396. 10.1016/j.ymthe.2018.08.017 - DOI - PMC - PubMed
-
- Boyer D.S., Schmidt-Erfurth U., van Lookeren Campagne M., Henry E.C., and Brittain C.. 2017. The Pathophysiology of Geographic Atrophy Secondary to Age-Related Macular Degeneration and the Complement Pathway as a Therapeutic Target. Retina. 37:819–835. 10.1097/IAE.0000000000001392 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

