The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35 is a thiolase superfamily member with a radically repurposed active site
- PMID: 31209106
- PMCID: PMC6682727
- DOI: 10.1074/jbc.RA119.008889
The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35 is a thiolase superfamily member with a radically repurposed active site
Abstract
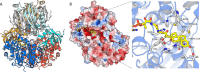
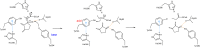
An aldolase from the bile acid-degrading actinobacterium Thermomonospora curvata catalyzes the C-C bond cleavage of an isopropyl-CoA side chain from the D-ring of the steroid metabolite 17-hydroxy-3-oxo-4-pregnene-20-carboxyl-CoA (17-HOPC-CoA). Like its homolog from Mycobacterium tuberculosis, the T. curvata aldolase is a protein complex of Ltp2 with a DUF35 domain derived from the C-terminal domain of a hydratase (ChsH2DUF35) that catalyzes the preceding step in the pathway. We determined the structure of the Ltp2-ChsH2DUF35 complex at 1.7 Å resolution using zinc-single anomalous diffraction. The enzyme adopts an αββα organization, with the two Ltp2 protomers forming a central dimer, and the two ChsH2DUF35 protomers being at the periphery. Docking experiments suggested that Ltp2 forms a tight complex with the hydratase but that each enzyme retains an independent CoA-binding site. Ltp2 adopted a fold similar to those in thiolases; however, instead of forming a deep tunnel, the Ltp2 active site formed an elongated cleft large enough to accommodate 17-HOPC-CoA. The active site lacked the two cysteines that served as the nucleophile and general base in thiolases and replaced a pair of oxyanion-hole histidine residues with Tyr-246 and Tyr-344. Phenylalanine replacement of either of these residues decreased aldolase catalytic activity at least 400-fold. On the basis of a 17-HOPC-CoA -docked model, we propose a catalytic mechanism where Tyr-294 acts as the general base abstracting a proton from the D-ring hydroxyl of 17-HOPC-CoA and Tyr-344 as the general acid that protonates the propionyl-CoA anion following C-C bond cleavage.
Keywords: C–C bond cleavage; actinobacteria; aldolase; bile acid; biodegradation; cholesterol; protein evolution; steroid; thiolase; β-oxidation.
© 2019 Aggett et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., and Eltis L. D. (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 10.1073/pnas.0605728104 - DOI - PMC - PubMed
-
- Dheda K., Gumbo T., Maartens G., Dooley K. E., McNerney R., Murray M., Furin J., Nardell E. A., London L., Lessem E., Theron G., van Helden P., Niemann S., Merker M., Dowdy D., et al. (2017) The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 5, 291–360 10.1016/S2213-2600(17)30079-6 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

