Intragenic antagonistic roles of protein and circRNA in tumorigenesis
- PMID: 31209250
- PMCID: PMC6796857
- DOI: 10.1038/s41422-019-0192-1
Intragenic antagonistic roles of protein and circRNA in tumorigenesis
Erratum in
-
Author Correction: Intragenic antagonistic roles of protein and circRNA in tumorigenesis.Cell Res. 2020 Feb;30(2):188. doi: 10.1038/s41422-019-0262-4. Cell Res. 2020. PMID: 31911670 Free PMC article.
Abstract
circRNAs arise from back splicing events during mRNA processing, and when deregulated can play an active role in cancer. Here we characterize a new circRNA (circPOK) encoded by the Zbtb7a gene (also kown as POKEMON, LRF) in the context of mesenchymal tumor progression. circPOK functions as a non-coding proto-oncogenic RNA independently and antithetically to its linear transcript counterpart, which acts as a tumor suppressor by encoding the Pokemon transcription factor. We find that circPOK regulates pro-proliferative and pro-angiogenic factors by co-activation of the ILF2/3 complex. Importantly, the expression of Pokemon protein and circRNA is aberrantly uncoupled in cancer through differential post-transcriptional regulation. Thus, we identify a novel type of genetic unit, the iRegulon, that yields biochemically distinct RNA products, circular and linear, with diverse and antithetical functions. Our findings further expand the cellular repertoire towards the control of normal biological outputs, while aberrant expression of such components may underlie disease pathogenesis including cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures
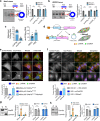


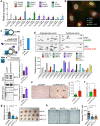

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

