Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia
- PMID: 31209280
- PMCID: PMC6893870
- DOI: 10.1038/s41375-019-0512-y
Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia
Abstract
Outcomes for patients with chronic myeloid leukemia (CML) have substantially improved due to advances in drug development and rational treatment intervention strategies. Despite these significant advances there are still unanswered questions on patient management regarding how to more reliably predict treatment failure at the time of diagnosis and how to select frontline tyrosine kinase inhibitor (TKI) therapy for optimal outcome. The BCR-ABL1 transcript level at diagnosis has no established prognostic impact and cannot guide frontline TKI selection. BCR-ABL1 mutations are detected in ~50% of TKI resistant patients but are rarely responsible for primary resistance. Other resistance mechanisms are largely uncharacterized and there are no other routine molecular testing strategies to facilitate the evaluation and further stratification of TKI resistance. Advances in next-generation sequencing technology has aided the management of a growing number of other malignancies, enabling the incorporation of somatic mutation profiles in diagnosis, classification, and prognostication. A largely unexplored area in CML research is whether expanded genomic analysis at diagnosis, resistance, and disease transformation can enhance patient management decisions, as has occurred for other cancers. The aim of this article is to review publications that reported mutated cancer-associated genes in CML patients at various disease phases. We discuss the frequency and type of such variants at initial diagnosis and at the time of treatment failure and transformation. Current limitations in the evaluation of mutants and recommendations for future reporting are outlined. The collective evaluation of mutational studies over more than a decade suggests a limited set of cancer-associated genes are indeed recurrently mutated in CML and some at a relatively high frequency. Genomic studies have the potential to lay the foundation for improved diagnostic risk classification according to clinical and genomic risk, and to enable more precise early identification of TKI resistance.
Conflict of interest statement
Conflict of Interest
SB: Member of the advisory board of Qiagen, Novartis and Bristol-Myers Squibb; Received honoraria from Qiagen, Novartis, Bristol-Myers Squibb and Cepheid. Research support from Novartis. CC: Honorarium from Novartis Oncology, Bristol-Myers Squibb, Korea Otsuka Pharmaceuticals, Chiltem International; Research funding from Bristol-Myers Squibb. BJD: Aileron Therapeutics, ALLCRON, Cepheid, Vivid Biosciences, Celgene, Gilead Sciences (inactive), Baxalta (inactive), Monojul (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, Third Coast Therapeutics, GRAIL (inactive), CTI BioPharma (inactive); Scientific Founder: MolecularMD (inactive, acquired by ICON); Board of Directors & Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license). TPH: Holds a consultancy role and has received research funding and honoraria from Novartis, Bristol-Myers Squibb and Ariad. Other authors declare no conflicts of interest.
Figures
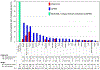



References
-
- Radich JP, Deininger M, Abboud CN, Altman JK, Berman E, Bhatia R, et al. Chronic Myeloid Leukemia, Version 1.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2018;16:1108–35. - PubMed
-
- Ashley EA. Towards precision medicine. Nature Reviews Genetics. 2016;17:507–22. - PubMed
-
- Sokal JE, Cox EB, Baccarani M, Tura S, Gomez GA, Robertson JE, et al. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood. 1984;63:789–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

