RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability
- PMID: 31209295
- PMCID: PMC6960015
- DOI: 10.1038/s41556-019-0337-y
RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability
Abstract
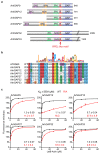
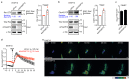
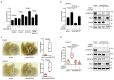
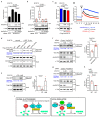
RPEL proteins, which contain the G-actin-binding RPEL motif, coordinate cytoskeletal processes with actin dynamics. We show that the ArhGAP12- and ArhGAP32-family GTPase-activating proteins (GAPs) are RPEL proteins. We determine the structure of the ArhGAP12/G-actin complex, and show that G-actin contacts the RPEL motif and GAP domain sequences. G-actin inhibits ArhGAP12 GAP activity, and this requires the G-actin contacts identified in the structure. In B16 melanoma cells, ArhGAP12 suppresses basal Rac and Cdc42 activity, F-actin assembly, invadopodia formation and experimental metastasis. In this setting, ArhGAP12 mutants defective for G-actin binding exhibit more effective downregulation of Rac GTP loading following HGF stimulation and enhanced inhibition of Rac-dependent processes, including invadopodia formation. Potentiation or disruption of the G-actin/ArhGAP12 interaction, by treatment with the actin-binding drugs latrunculin B or cytochalasin D, has corresponding effects on Rac GTP loading. The interaction of G-actin with RPEL-family rhoGAPs thus provides a negative feedback loop that couples Rac activity to actin dynamics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Stimulation of fascin spikes by thrombospondin-1 is mediated by the GTPases Rac and Cdc42.J Cell Biol. 2000 Aug 21;150(4):807-22. doi: 10.1083/jcb.150.4.807. J Cell Biol. 2000. PMID: 10953005 Free PMC article.
-
Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation.Plant Physiol. 2000 Dec;124(4):1625-36. doi: 10.1104/pp.124.4.1625. Plant Physiol. 2000. PMID: 11115880 Free PMC article.
-
GLUT4 vesicle recruitment and fusion are differentially regulated by Rac, AS160, and Rab8A in muscle cells.J Biol Chem. 2008 Oct 3;283(40):27208-19. doi: 10.1074/jbc.M804282200. Epub 2008 Jul 23. J Biol Chem. 2008. PMID: 18650435
-
ArhGAP12 plays dual roles in Stabilin-2 mediated efferocytosis: Regulates Rac1 basal activity and spatiotemporally turns off the Rac1 to orchestrate phagosome maturation.Biochim Biophys Acta Mol Cell Res. 2019 Oct;1866(10):1595-1607. doi: 10.1016/j.bbamcr.2019.07.003. Epub 2019 Jul 10. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 31301364
-
Ras-related GTPases and the cytoskeleton.Mol Biol Cell. 1992 May;3(5):475-9. doi: 10.1091/mbc.3.5.475. Mol Biol Cell. 1992. PMID: 1611153 Free PMC article. Review.
Cited by
-
Rac-maninoff and Rho-vel: The symphony of Rho-GTPase signaling at excitatory synapses.Small GTPases. 2022 Jan;13(1):14-47. doi: 10.1080/21541248.2021.1885264. Epub 2021 May 6. Small GTPases. 2022. PMID: 33955328 Free PMC article.
-
Patterning of the cell cortex by Rho GTPases.Nat Rev Mol Cell Biol. 2024 Apr;25(4):290-308. doi: 10.1038/s41580-023-00682-z. Epub 2024 Jan 3. Nat Rev Mol Cell Biol. 2024. PMID: 38172611 Review.
-
Macrophages form dendrite-like pseudopods to enhance bacterial ingestion.EMBO J. 2025 Jul 28. doi: 10.1038/s44318-025-00515-z. Online ahead of print. EMBO J. 2025. PMID: 40721684
-
Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.Elife. 2020 Sep 25;9:e61509. doi: 10.7554/eLife.61509. Elife. 2020. PMID: 32975518 Free PMC article.
-
Hepatocellular carcinoma resistance to tyrosine kinase inhibitors: Current status and perspectives.World J Gastrointest Oncol. 2025 Apr 15;17(4):101528. doi: 10.4251/wjgo.v17.i4.101528. World J Gastrointest Oncol. 2025. PMID: 40235904 Free PMC article.
References
-
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. - PubMed
-
- Przybyla L, Muncie JM, Weaver VM. Mechanical Control of Epithelial-to-Mesenchymal Transitions in Development and Cancer. Annu Rev Cell Dev Biol. 2016;32:527–554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

