Inferring protein 3D structure from deep mutation scans
- PMID: 31209393
- PMCID: PMC7295002
- DOI: 10.1038/s41588-019-0432-9
Inferring protein 3D structure from deep mutation scans
Abstract
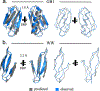
We describe an experimental method of three-dimensional (3D) structure determination that exploits the increasing ease of high-throughput mutational scans. Inspired by the success of using natural, evolutionary sequence covariation to compute protein and RNA folds, we explored whether 'laboratory', synthetic sequence variation might also yield 3D structures. We analyzed five large-scale mutational scans and discovered that the pairs of residues with the largest positive epistasis in the experiments are sufficient to determine the 3D fold. We show that the strongest epistatic pairings from genetic screens of three proteins, a ribozyme and a protein interaction reveal 3D contacts within and between macromolecules. Using these experimental epistatic pairs, we compute ab initio folds for a GB1 domain (within 1.8 Å of the crystal structure) and a WW domain (2.1 Å). We propose strategies that reduce the number of mutants needed for contact prediction, suggesting that genomics-based techniques can efficiently predict 3D structure.
Figures



References
Methods-only References
-
- Bonneau RT,J; Chivian D; Rohl C; Strauss C; Baker D. Rosetta in CASP4: progress in ab initio protein structure prediction. PROTEINS: Structure, Function, and Genetics 5, 119–126 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

