Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues
- PMID: 31209406
- PMCID: PMC6685551
- DOI: 10.1038/s41590-019-0425-y
Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues
Erratum in
-
Publisher Correction: Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues.Nat Immunol. 2019 Oct;20(10):1405. doi: 10.1038/s41590-019-0474-2. Nat Immunol. 2019. PMID: 31388149
Abstract
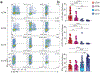
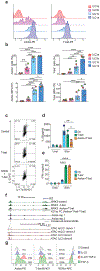
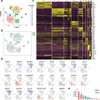
Innate lymphoid cells (ILCs) are tissue-resident lymphocytes categorized on the basis of their core regulatory programs and the expression of signature cytokines. Human ILC3s that produce the cytokine interleukin-22 convert into ILC1-like cells that produce interferon-γ in vitro, but whether this conversion occurs in vivo remains unclear. In the present study we found that ILC3s and ILC1s in human tonsils represented the ends of a spectrum that included additional discrete subsets. RNA velocity analysis identified an intermediate ILC3-ILC1 cluster, which had strong directionality toward ILC1s. In humanized mice, the acquisition of ILC1 features by ILC3s showed tissue dependency. Chromatin studies indicated that the transcription factors Aiolos and T-bet cooperated to repress regulatory elements active in ILC3s. A transitional ILC3-ILC1 population was also detected in the human intestine. We conclude that ILC3s undergo conversion into ILC1-like cells in human tissues in vivo, and that tissue factors and Aiolos were required for this process.
Conflict of interest statement
Competing Interests
R.G., S.Z., W.G., J.S., L.-L.L., M.B. and S.A.J. are Pfizer employees. M. Colonna received funding from Pfizer to study ILC biology in Inflammatory Bowel Disease. The other authors declare no conflicts of interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA188286/CA/NCI NIH HHS/United States
- P30 AR073752/AR/NIAMS NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- P01 AI106697/AI/NIAID NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R01 DE025884/DE/NIDCR NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- U01 AI095542/AI/NIAID NIH HHS/United States
- K99 DK118110/DK/NIDDK NIH HHS/United States
- R01 AI134035/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

