Supporting non-target identification by adding hydrogen deuterium exchange MS/MS capabilities to MetFrag
- PMID: 31209548
- PMCID: PMC6611743
- DOI: 10.1007/s00216-019-01885-0
Supporting non-target identification by adding hydrogen deuterium exchange MS/MS capabilities to MetFrag
Abstract
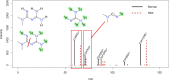
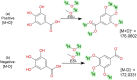
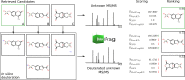
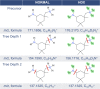
Liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) is increasingly popular for the non-targeted exploration of complex samples, where tandem mass spectrometry (MS/MS) is used to characterize the structure of unknown compounds. However, mass spectra do not always contain sufficient information to unequivocally identify the correct structure. This study investigated how much additional information can be gained using hydrogen deuterium exchange (HDX) experiments. The exchange of "easily exchangeable" hydrogen atoms (connected to heteroatoms), with predominantly [M+D]+ ions in positive mode and [M-D]- in negative mode was observed. To enable high-throughput processing, new scoring terms were incorporated into the in silico fragmenter MetFrag. These were initially developed on small datasets and then tested on 762 compounds of environmental interest. Pairs of spectra (normal and deuterated) were found for 593 of these substances (506 positive mode, 155 negative mode spectra). The new scoring terms resulted in 29 additional correct identifications (78 vs 49) for positive mode and an increase in top 10 rankings from 80 to 106 in negative mode. Compounds with dual functionality (polar head group, long apolar tail) exhibited dramatic retention time (RT) shifts of up to several minutes, compared with an average 0.04 min RT shift. For a smaller dataset of 80 metabolites, top 10 rankings improved from 13 to 24 (positive mode, 57 spectra) and from 14 to 31 (negative mode, 63 spectra) when including HDX information. The results of standard measurements were confirmed using targets and tentatively identified surfactant species in an environmental sample collected from the river Danube near Novi Sad (Serbia). The changes to MetFrag have been integrated into the command line version available at http://c-ruttkies.github.io/MetFrag and all resulting spectra and compounds are available in online resources and in the Electronic Supplementary Material (ESM). Graphical abstract.
Keywords: Compound identification; High-resolution mass spectrometry; Hydrogen deuterium exchange; In silico fragmentation; Metabolomics; Structure elucidation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

