Growth of Dehalococcoides spp. and increased abundance of reductive dehalogenase genes in anaerobic PCB-contaminated sediment microcosms
- PMID: 31209752
- PMCID: PMC6918016
- DOI: 10.1007/s11356-019-05571-7
Growth of Dehalococcoides spp. and increased abundance of reductive dehalogenase genes in anaerobic PCB-contaminated sediment microcosms
Abstract
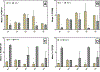
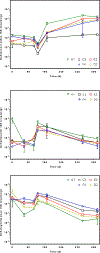
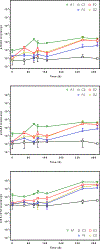
Polychlorinated biphenyls (PCBs) contaminate 19% of US Superfund sites and represent a serious risk to human and environmental health. One promising strategy to remediate PCB-contaminated sediments utilizes organohalide-respiring bacteria (OHRB) that dechlorinate PCBs.However, functional genes that act as biomarkers for PCB dechlorination processes (i.e., reductive dehalogenase genes) are poorly understood. Here, we developed anaerobic sediment microcosms that harbor an OHRB community dominated by the genus Dehalococcoides. During the 430-day microcosm incubation, Dehalococcoides 16S rRNA sequences increased two orders of magnitude to 107 copies/g of sediment, and at the same time, PCB118 decreased by as much as 70%. In addition, the OHRB community dechlorinated a range of penta- and tetra-chlorinated PCB congeners including PCBs 66, 70 + 74 + 76, 95, 90 + 101, and PCB110 without exogenous electron donor. We quantified candidate reductive dehalogenase (RDase) genes over a 430-day incubation period and found rd14, a reductive dehalogenase that belongs to Dehalococcoides mccartyi strain CG5, was enriched to 107 copies/g of sediment. At the same time, pcbA5 was enriched to only 105 copies/g of sediment. A survey for additional RDase genes revealed sequences similar to strain CG5's rd4 and rd8. In addition to demonstrating the PCB dechlorination potential of native microbial communities in contaminated freshwater sediments, our results suggest candidate functional genes with previously unexplored potential could serve as biomarkers of PCB dechlorination processes.
Keywords: Dehalococcoides mccartyi; PCBs; Polychlorinated biphenyl; Reductive dechlorination; Reductive dehalogenase genes; Sediment microcosms.
Figures



References
-
- Abraham W-R, Nogales B, Golyshin PN, Pieper DH, Timmis KN (2002): Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol 5, 246–253 - PubMed
-
- Anezaki K, Nakano T (2014): Concentration levels and congener profiles of polychlorinated biphenyls, pentachlorobenzene, and hexachlorobenzene in commercial pigments. Environ Sci Poll Res 21, 998–1009 - PubMed
-
- Chen C, He J (2018): Strategy for the rapid dechlorination of Polychlorinated Biphenyls (PCBs) by Dehalococcoides mccartyi strains. Environ. Sci. Technol 52, 13854–13862 - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

