Transcriptomic correlates of electrophysiological and morphological diversity within and across excitatory and inhibitory neuron classes
- PMID: 31211786
- PMCID: PMC6599125
- DOI: 10.1371/journal.pcbi.1007113
Transcriptomic correlates of electrophysiological and morphological diversity within and across excitatory and inhibitory neuron classes
Abstract
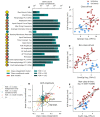
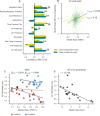
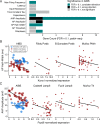
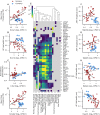
In order to further our understanding of how gene expression contributes to key functional properties of neurons, we combined publicly accessible gene expression, electrophysiology, and morphology measurements to identify cross-cell type correlations between these data modalities. Building on our previous work using a similar approach, we distinguished between correlations which were "class-driven," meaning those that could be explained by differences between excitatory and inhibitory cell classes, and those that reflected graded phenotypic differences within classes. Taking cell class identity into account increased the degree to which our results replicated in an independent dataset as well as their correspondence with known modes of ion channel function based on the literature. We also found a smaller set of genes whose relationships to electrophysiological or morphological properties appear to be specific to either excitatory or inhibitory cell types. Next, using data from PatchSeq experiments, allowing simultaneous single-cell characterization of gene expression and electrophysiology, we found that some of the gene-property correlations observed across cell types were further predictive of within-cell type heterogeneity. In summary, we have identified a number of relationships between gene expression, electrophysiology, and morphology that provide testable hypotheses for future studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gouwens NW, Sorensen SA, Berg J, Lee C, Jarsky T, Ting J, et al. Classification of electrophysiological and morphological types in mouse visual cortex. 2018;Preprint. Available from: https://www.biorxiv.org/content/biorxiv/early/2018/07/17/368456.full.pdf - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

