Establishment of a Cell Culture Model of Persistent Flaviviral Infection: Usutu Virus Shows Sustained Replication during Passages and Resistance to Extinction by Antiviral Nucleosides
- PMID: 31212939
- PMCID: PMC6630443
- DOI: 10.3390/v11060560
Establishment of a Cell Culture Model of Persistent Flaviviral Infection: Usutu Virus Shows Sustained Replication during Passages and Resistance to Extinction by Antiviral Nucleosides
Abstract
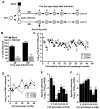
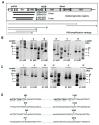
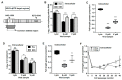
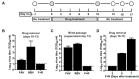
Chronic viral disease constitutes a major global health problem, with several hundred million people affected and an associated elevated number of deaths. An increasing number of disorders caused by human flaviviruses are related to their capacity to establish a persistent infection. Here we show that Usutu virus (USUV), an emerging zoonotic flavivirus linked to sporadic neurologic disease in humans, can establish a persistent infection in cell culture. Two independent lineages of Vero cells surviving USUV lytic infection were cultured over 82 days (41 cell transfers) without any apparent cytopathology crisis associated. We found elevated titers in the supernatant of these cells, with modest fluctuations during passages but no overall tendency towards increased or decreased infectivity. In addition to full-length genomes, viral RNA isolated from these cells at passage 40 revealed the presence of defective genomes, containing different deletions at the 5' end. These truncated transcripts were all predicted to encode shorter polyprotein products lacking membrane and envelope structural proteins, and most of non-structural protein 1. Treatment with different broad-range antiviral nucleosides revealed that USUV is sensitive to these compounds in the context of a persistent infection, in agreement with previous observations during lytic infections. The exposure of infected cells to prolonged treatment (10 days) with favipiravir and/or ribavirin resulted in the complete clearance of infectivity in the cellular supernatants (decrease of ~5 log10 in virus titers and RNA levels), although modest changes in intracellular viral RNA levels were recorded (<2 log10 decrease). Drug withdrawal after treatment day 10 resulted in a relapse in virus titers. These results encourage the use of persistently-infected cultures as a surrogate system in the identification of improved antivirals against flaviviral chronic disease.
Keywords: antiviral therapies; chronic viral infection; defective viral genomes; emerging arboviruses; lethal mutagenesis.
Conflict of interest statement
We declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Extinction of Zika Virus and Usutu Virus by Lethal Mutagenesis Reveals Different Patterns of Sensitivity to Three Mutagenic Drugs.Antimicrob Agents Chemother. 2018 Aug 27;62(9):e00380-18. doi: 10.1128/AAC.00380-18. Print 2018 Sep. Antimicrob Agents Chemother. 2018. PMID: 29914957 Free PMC article.
-
Favipiravir inhibits in vitro Usutu virus replication and delays disease progression in an infection model in mice.Antiviral Res. 2018 Dec;160:137-142. doi: 10.1016/j.antiviral.2018.10.026. Epub 2018 Oct 29. Antiviral Res. 2018. PMID: 30385306
-
Favipiravir elicits antiviral mutagenesis during virus replication in vivo.Elife. 2014 Oct 21;3:e03679. doi: 10.7554/eLife.03679. Elife. 2014. PMID: 25333492 Free PMC article.
-
Molecular Insights into the Flavivirus Replication Complex.Viruses. 2021 May 21;13(6):956. doi: 10.3390/v13060956. Viruses. 2021. PMID: 34064113 Free PMC article. Review.
-
The importance of viral and cellular factors on flavivirus entry.Curr Opin Virol. 2021 Aug;49:164-175. doi: 10.1016/j.coviro.2021.05.001. Epub 2021 Jun 23. Curr Opin Virol. 2021. PMID: 34171540 Review.
Cited by
-
Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus.Int J Mol Sci. 2021 Apr 19;22(8):4218. doi: 10.3390/ijms22084218. Int J Mol Sci. 2021. PMID: 33921710 Free PMC article.
-
New Insights into the Susceptibility of Immunocompetent Mice to Usutu Virus.Viruses. 2020 Feb 8;12(2):189. doi: 10.3390/v12020189. Viruses. 2020. PMID: 32046265 Free PMC article.
-
Relevance of oxidative stress in inhibition of eIF2 alpha phosphorylation and stress granules formation during Usutu virus infection.PLoS Negl Trop Dis. 2021 Jan 25;15(1):e0009072. doi: 10.1371/journal.pntd.0009072. eCollection 2021 Jan. PLoS Negl Trop Dis. 2021. PMID: 33493202 Free PMC article.
-
Virus-induced brain pathology and the neuroinflammation-inflammation continuum: the neurochemists view.J Neural Transm (Vienna). 2024 Dec;131(12):1429-1453. doi: 10.1007/s00702-023-02723-5. Epub 2024 Jan 23. J Neural Transm (Vienna). 2024. PMID: 38261034 Free PMC article. Review.
-
Ubiquitin-like modifier-activating enzyme 1 interacts with Zika virus NS5 and promotes viral replication in the infected cell.J Gen Virol. 2025 Jan;106(1):002063. doi: 10.1099/jgv.0.002063. J Gen Virol. 2025. PMID: 39773572 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

