Analysis of Hematological Traits in Polled Yak by Genome-Wide Association Studies Using Individual SNPs and Haplotypes
- PMID: 31212963
- PMCID: PMC6627507
- DOI: 10.3390/genes10060463
Analysis of Hematological Traits in Polled Yak by Genome-Wide Association Studies Using Individual SNPs and Haplotypes
Abstract
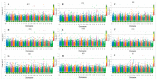
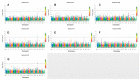
Yak (Bos grunniens) is an important domestic animal living in high-altitude plateaus. Due to inadequate disease prevention, each year, the yak industry suffers significant economic losses. The identification of causal genes that affect blood- and immunity-related cells could provide preliminary reference guidelines for the prevention of diseases in the population of yaks. The genome-wide association studies (GWASs) utilizing a single-marker or haplotype method were employed to analyze 15 hematological traits in the genome of 315 unrelated yaks. Single-marker GWASs identified a total of 43 significant SNPs, including 35 suggestive and eight genome-wide significant SNPs, associated with nine traits. Haplotype analysis detected nine significant haplotype blocks, including two genome-wide and seven suggestive blocks, associated with seven traits. The study provides data on the genetic variability of hematological traits in the yak. Five essential genes (GPLD1, EDNRA,APOB, HIST1H1E, and HIST1H2BI) were identified, which affect the HCT, HGB, RBC, PDW, PLT, and RDWSD traits and can serve as candidate genes for regulating hematological traits. The results provide a valuable reference to be used in the analysis of blood properties and immune diseases in the yak.
Keywords: haplotype analysis; hematological traits; polled yak; single-marker GWAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Genome-Wide Association Study Using Individual Single-Nucleotide Polymorphisms and Haplotypes for Erythrocyte Traits in Alpine Merino Sheep.Front Genet. 2020 Jul 31;11:848. doi: 10.3389/fgene.2020.00848. eCollection 2020. Front Genet. 2020. PMID: 32849829 Free PMC article.
-
Associations of single nucleotide polymorphisms in candidate genes with the polled trait in Datong domestic yaks.Anim Genet. 2014 Feb;45(1):138-41. doi: 10.1111/age.12081. Epub 2013 Aug 27. Anim Genet. 2014. PMID: 24033474
-
Genetic diversities of MT-ND1 and MT-ND2 genes are associated with high-altitude adaptation in yak.Mitochondrial DNA A DNA Mapp Seq Anal. 2018 Apr;29(3):485-494. doi: 10.1080/24701394.2017.1307976. Epub 2017 Apr 1. Mitochondrial DNA A DNA Mapp Seq Anal. 2018. PMID: 28366030
-
Genome-wide association study of porcine hematological parameters in a Large White × Minzhu F2 resource population.Int J Biol Sci. 2012;8(6):870-81. doi: 10.7150/ijbs.4027. Epub 2012 Jun 15. Int J Biol Sci. 2012. PMID: 22745577 Free PMC article.
-
Mapping genes for complex traits in domestic animals and their use in breeding programmes.Nat Rev Genet. 2009 Jun;10(6):381-91. doi: 10.1038/nrg2575. Nat Rev Genet. 2009. PMID: 19448663 Review.
Cited by
-
Genome-Wide Association Study Using Individual Single-Nucleotide Polymorphisms and Haplotypes for Erythrocyte Traits in Alpine Merino Sheep.Front Genet. 2020 Jul 31;11:848. doi: 10.3389/fgene.2020.00848. eCollection 2020. Front Genet. 2020. PMID: 32849829 Free PMC article.
-
Genome-Wide Association Study Identifies New Candidate Markers for Somatic Cells Score in a Local Dairy Sheep.Front Genet. 2021 Mar 22;12:643531. doi: 10.3389/fgene.2021.643531. eCollection 2021. Front Genet. 2021. PMID: 33828586 Free PMC article.
-
The Pan-Immune Inflammation Value at Admission Predicts Postoperative in-hospital Mortality in Patients with Acute Type A Aortic Dissection.J Inflamm Res. 2024 Aug 5;17:5223-5234. doi: 10.2147/JIR.S468017. eCollection 2024. J Inflamm Res. 2024. PMID: 39131211 Free PMC article.
-
A Genome-Wide Association Study Identifies Quantitative Trait Loci Affecting Hematological Traits in Camelus bactrianus.Animals (Basel). 2020 Jan 7;10(1):96. doi: 10.3390/ani10010096. Animals (Basel). 2020. PMID: 31936121 Free PMC article.
-
Weighted Single-Step Genome-Wide Association Study Uncovers Known and Novel Candidate Genomic Regions for Milk Production Traits and Somatic Cell Score in Valle del Belice Dairy Sheep.Animals (Basel). 2022 Apr 29;12(9):1155. doi: 10.3390/ani12091155. Animals (Basel). 2022. PMID: 35565582 Free PMC article.
References
-
- Wang Z., Shen X., Liu B., Su J., Yonezawa T., Yu Y., Guo S., Ho S.Y., Vila C., Hasegawa M. Phylogeographical analyses of domestic and wild yaks based on mitochondrial DNA: New data and reappraisal. J. Biogeogr. 2010;37:2332–2344. doi: 10.1111/j.1365-2699.2010.02379.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

