High-throughput analysis revealed mutations' diverging effects on SMN1 exon 7 splicing
- PMID: 31213135
- PMCID: PMC6779402
- DOI: 10.1080/15476286.2019.1630796
High-throughput analysis revealed mutations' diverging effects on SMN1 exon 7 splicing
Abstract
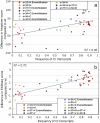
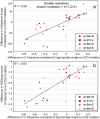
Splicing-affecting mutations can disrupt gene function by altering the transcript assembly. To ascertain splicing dysregulation principles, we modified a minigene assay for the parallel high-throughput evaluation of different mutations by next-generation sequencing. In our model system, all exonic and six intronic positions of the SMN1 gene's exon 7 were mutated to all possible nucleotide variants, which amounted to 180 unique single-nucleotide mutants and 470 double mutants. The mutations resulted in a wide range of splicing aberrations. Exonic splicing-affecting mutations resulted either in substantial exon skipping, supposedly driven by predicted exonic splicing silencer or cryptic donor splice site (5'ss) and de novo 5'ss strengthening and use. On the other hand, a single disruption of exonic splicing enhancer was not sufficient to cause major exon skipping, suggesting these elements can be substituted during exon recognition. While disrupting the acceptor splice site led only to exon skipping, some 5'ss mutations potentiated the use of three different cryptic 5'ss. Generally, single mutations supporting cryptic 5'ss use displayed better pre-mRNA/U1 snRNA duplex stability and increased splicing regulatory element strength across the original 5'ss. Analyzing double mutants supported the predominating splicing regulatory elements' effect, but U1 snRNA binding could contribute to the global balance of splicing isoforms. Based on these findings, we suggest that creating a new splicing enhancer across the mutated 5'ss can be one of the main factors driving cryptic 5'ss use.
Keywords: 5′ss; SMN1; U1 snRNA; cryptic splice sites; splicing-affecting mutation.
Figures
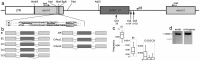

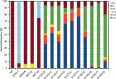
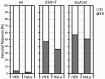
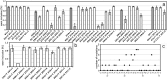

Similar articles
-
Regulation of a strong F9 cryptic 5'ss by intrinsic elements and by combination of tailored U1snRNAs with antisense oligonucleotides.Hum Mol Genet. 2015 Sep 1;24(17):4809-16. doi: 10.1093/hmg/ddv205. Epub 2015 Jun 10. Hum Mol Genet. 2015. PMID: 26063760 Free PMC article.
-
How to Design U1 snRNA Molecules for Splicing Rescue.Methods Mol Biol. 2022;2434:89-102. doi: 10.1007/978-1-0716-2010-6_5. Methods Mol Biol. 2022. PMID: 35213011 Free PMC article.
-
An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects.Hum Mol Genet. 2012 Jun 1;21(11):2389-98. doi: 10.1093/hmg/dds045. Epub 2012 Feb 23. Hum Mol Genet. 2012. PMID: 22362925 Free PMC article.
-
A novel role of U1 snRNP: Splice site selection from a distance.Biochim Biophys Acta Gene Regul Mech. 2019 Jun;1862(6):634-642. doi: 10.1016/j.bbagrm.2019.04.004. Epub 2019 Apr 28. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31042550 Free PMC article. Review.
-
Pick one, but be quick: 5' splice sites and the problems of too many choices.Genes Dev. 2013 Jan 15;27(2):129-44. doi: 10.1101/gad.209759.112. Genes Dev. 2013. PMID: 23348838 Free PMC article. Review.
Cited by
-
Mutations primarily alter the inclusion of alternatively spliced exons.Elife. 2020 Oct 28;9:e59959. doi: 10.7554/eLife.59959. Elife. 2020. PMID: 33112234 Free PMC article.
-
Splicing analysis of STAT3 tandem donor suggests non-canonical binding registers for U1 and U6 snRNAs.Nucleic Acids Res. 2024 Jun 10;52(10):5959-5974. doi: 10.1093/nar/gkae147. Nucleic Acids Res. 2024. PMID: 38426935 Free PMC article.
-
Splicing in the Diagnosis of Rare Disease: Advances and Challenges.Front Genet. 2021 Jul 1;12:689892. doi: 10.3389/fgene.2021.689892. eCollection 2021. Front Genet. 2021. PMID: 34276790 Free PMC article. Review.
-
Insulin-like growth factor 2 mRNA-binding protein 2-regulated alternative splicing of nuclear factor 1 C-type causes excessive granulosa cell proliferation in polycystic ovary syndrome.Cell Prolif. 2022 Apr;55(4):e13216. doi: 10.1111/cpr.13216. Epub 2022 Mar 16. Cell Prolif. 2022. PMID: 35293050 Free PMC article.
-
Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects.Nat Rev Genet. 2023 Apr;24(4):251-269. doi: 10.1038/s41576-022-00556-8. Epub 2022 Dec 16. Nat Rev Genet. 2023. PMID: 36526860 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
