RNA-protein interactions: disorder, moonlighting and junk contribute to eukaryotic complexity
- PMID: 31213136
- PMCID: PMC6597761
- DOI: 10.1098/rsob.190096
RNA-protein interactions: disorder, moonlighting and junk contribute to eukaryotic complexity
Abstract
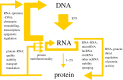
RNA-protein interactions are crucial for most biological processes in all organisms. However, it appears that the complexity of RNA-based regulation increases with the complexity of the organism, creating additional regulatory circuits, the scope of which is only now being revealed. It is becoming apparent that previously unappreciated features, such as disordered structural regions in proteins or non-coding regions in DNA leading to higher plasticity and pliability in RNA-protein complexes, are in fact essential for complex, precise and fine-tuned regulation. This review addresses the issue of the role of RNA-protein interactions in generating eukaryotic complexity, focusing on the newly characterized disordered RNA-binding motifs, moonlighting of metabolic enzymes, RNA-binding proteins interactions with different RNA species and their participation in regulatory networks of higher order.
Keywords: RNA-binding proteins; eukaryotic complexity; intrinsically disordered proteins; non-coding RNA; protein moonlighting.
Conflict of interest statement
We have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

