Human FAM173A is a mitochondrial lysine-specific methyltransferase that targets adenine nucleotide translocase and affects mitochondrial respiration
- PMID: 31213526
- PMCID: PMC6682728
- DOI: 10.1074/jbc.RA119.009045
Human FAM173A is a mitochondrial lysine-specific methyltransferase that targets adenine nucleotide translocase and affects mitochondrial respiration
Abstract
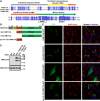
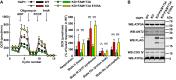
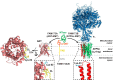
Lysine methylation is a common posttranslational modification of nuclear and cytoplasmic proteins but is also present in mitochondria. The human protein denoted "family with sequence similarity 173 member B" (FAM173B) was recently uncovered as a mitochondrial lysine (K)-specific methyltransferase (KMT) targeting the c-subunit of mitochondrial ATP synthase (ATPSc), and was therefore renamed ATPSc-KMT. We here set out to investigate the biochemical function of its yet uncharacterized paralogue FAM173A. We demonstrate that FAM173A localizes to mitochondria, mediated by a noncanonical targeting sequence that is partially retained in the mature protein. Immunoblotting analysis using methyllysine-specific antibodies revealed that FAM173A knock-out (KO) abrogates lysine methylation of a single mitochondrial protein in human cells. Mass spectrometry analysis identified this protein as adenine nucleotide translocase (ANT), represented by two highly similar isoforms ANT2 and ANT3. We found that methylation occurs at Lys-52 of ANT, which was previously reported to be trimethylated. Complementation of KO cells with WT or enzyme-dead FAM173A indicated that the enzymatic activity of FAM173A is required for ANT methylation at Lys-52 to occur. Both in human cells and in rat organs, Lys-52 was exclusively trimethylated, indicating that this modification is constitutive, rather than regulatory and dynamic. Moreover, FAM173A-deficient cells displayed increased mitochondrial respiration compared with FAM173A-proficient cells. In summary, we demonstrate that FAM173A is the long-sought KMT responsible for ANT methylation at Lys-52, and point out the functional significance of Lys-52 methylation in ANT. Based on the established naming nomenclature for KMTs, we propose to rename FAM173A to ANT-KMT (gene name ANTKMT).
Keywords: ADP/ATP carrier; ADP/ATP translocase; ANT-KMT; FAM173A; KMT; adenine nucleotide translocase; bioenergetics; cardiolipin; lysine methylation; membrane enzyme; methyltransferase; mitochondria; mitochondrial transport; nucleoside/nucleotide transport; posttranslational modification (PTM); protein methylation; proteomics; solute carrier protein; transmembrane domain; transmembrane protein.
© 2019 Małecki et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

