Integration of Fungus-Specific CandA-C1 into a Trimeric CandA Complex Allowed Splitting of the Gene for the Conserved Receptor Exchange Factor of CullinA E3 Ubiquitin Ligases in Aspergilli
- PMID: 31213557
- PMCID: PMC6581859
- DOI: 10.1128/mBio.01094-19
Integration of Fungus-Specific CandA-C1 into a Trimeric CandA Complex Allowed Splitting of the Gene for the Conserved Receptor Exchange Factor of CullinA E3 Ubiquitin Ligases in Aspergilli
Abstract
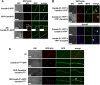
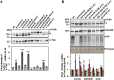
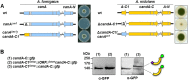
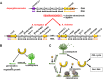
E3 cullin-RING ubiquitin ligase (CRL) complexes recognize specific substrates and are activated by covalent modification with ubiquitin-like Nedd8. Deneddylation inactivates CRLs and allows Cand1/A to bind and exchange substrate recognition subunits. Human as well as most fungi possess a single gene for the receptor exchange factor Cand1, which is split and rearranged in aspergilli into two genes for separate proteins. Aspergillus nidulans CandA-N blocks the neddylation site, and CandA-C inhibits the interaction to the adaptor/substrate receptor subunits similar to the respective N-terminal and C-terminal parts of single Cand1. The pathogen Aspergillus fumigatus and related species express a CandA-C with a 190-amino-acid N-terminal extension domain encoded by an additional exon. This extension corresponds in most aspergilli, including A. nidulans, to a gene directly upstream of candA-C encoding a 20-kDa protein without human counterpart. This protein was named CandA-C1, because it is also required for the cellular deneddylation/neddylation cycle and can form a trimeric nuclear complex with CandA-C and CandA-N. CandA-C and CandA-N are required for asexual and sexual development and control a distinct secondary metabolism. CandA-C1 and the corresponding domain of A. fumigatus control spore germination, vegetative growth, and the repression of additional secondary metabolites. This suggests that the dissection of the conserved Cand1-encoding gene within the genome of aspergilli was possible because it allowed the integration of a fungus-specific protein required for growth into the CandA complex in two different gene set versions, which might provide an advantage in evolution.IMPORTANCEAspergillus species are important for biotechnological applications, like the production of citric acid or antibacterial agents. Aspergilli can cause food contamination or invasive aspergillosis to immunocompromised humans or animals. Specific treatment is difficult due to limited drug targets and emerging resistances. The CandA complex regulates, as a receptor exchange factor, the activity and substrate variability of the ubiquitin labeling machinery for 26S proteasome-mediated protein degradation. Only Aspergillus species encode at least two proteins that form a CandA complex. This study shows that Aspergillus species had to integrate a third component into the CandA receptor exchange factor complex that is unique to aspergilli and required for vegetative growth, sexual reproduction, and activation of the ubiquitin labeling machinery. These features have interesting implications for the evolution of protein complexes and could make CandA-C1 an interesting candidate for target-specific drug design to control fungal growth without affecting the human ubiquitin-proteasome system.
Keywords: Aspergillus fumigatus; Aspergillus nidulans; COP9 signalosome; Cand1; Cullin-RING ubiquitin ligase; Nedd8; asexual development; protein complex; protein degradation; secondary metabolism; sexual development; spore germination.
Copyright © 2019 Köhler et al.
Figures





Similar articles
-
The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development.Mol Microbiol. 2012 Mar;83(6):1162-77. doi: 10.1111/j.1365-2958.2012.07999.x. Epub 2012 Feb 22. Mol Microbiol. 2012. PMID: 22329854
-
Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site.Mol Biol Cell. 2011 Jan 1;22(1):153-64. doi: 10.1091/mbc.E10-08-0732. Epub 2010 Nov 30. Mol Biol Cell. 2011. PMID: 21119001 Free PMC article.
-
The COP9 signalosome inhibits Cullin-RING E3 ubiquitin ligases independently of its deneddylase activity.Fly (Austin). 2018;12(2):118-126. doi: 10.1080/19336934.2018.1429858. Epub 2018 Feb 9. Fly (Austin). 2018. PMID: 29355077 Free PMC article.
-
Protection of cullin-RING E3 ligases by CSN-UBP12.Trends Cell Biol. 2006 Jul;16(7):362-9. doi: 10.1016/j.tcb.2006.05.001. Epub 2006 Jun 9. Trends Cell Biol. 2006. PMID: 16762551 Review.
-
Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases.Antioxid Redox Signal. 2010 Dec 1;13(11):1699-712. doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14. Antioxid Redox Signal. 2010. PMID: 20486766 Free PMC article. Review.
Cited by
-
Regulators of the Asexual Life Cycle of Aspergillus nidulans.Cells. 2023 Jun 4;12(11):1544. doi: 10.3390/cells12111544. Cells. 2023. PMID: 37296664 Free PMC article. Review.
-
The Third International Symposium on Fungal Stress - ISFUS.Fungal Biol. 2020 May;124(5):235-252. doi: 10.1016/j.funbio.2020.02.007. Epub 2020 Feb 24. Fungal Biol. 2020. PMID: 32389286 Free PMC article.
-
Post-translational modifications drive secondary metabolite biosynthesis in Aspergillus: a review.Environ Microbiol. 2022 Jul;24(7):2857-2881. doi: 10.1111/1462-2920.16034. Epub 2022 May 30. Environ Microbiol. 2022. PMID: 35645150 Free PMC article. Review.
-
The velvet protein Vel1 controls initial plant root colonization and conidia formation for xylem distribution in Verticillium wilt.PLoS Genet. 2021 Mar 15;17(3):e1009434. doi: 10.1371/journal.pgen.1009434. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33720931 Free PMC article.
-
Fungal COP9 signalosome assembly requires connection of two trimeric intermediates for integration of intrinsic deneddylase.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2305049120. doi: 10.1073/pnas.2305049120. Epub 2023 Aug 21. Proc Natl Acad Sci U S A. 2023. PMID: 37603767 Free PMC article.
References
-
- Riquelme M, Aguirre J, Bartnicki-García S, Braus GH, Feldbrügge M, Fleig U, Hansberg W, Herrera-Estrella A, Kämper J, Kück U, Mouriño-Pérez RR, Takeshita N, Fischer R. 2018. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol Mol Biol Rev 82:e00068-17. doi:10.1128/MMBR.00068-17. - DOI - PMC - PubMed
-
- de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram Ö, Benocci T, Braus-Stromeyer SA, Caldana C, Cánovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RA, Damásio AR, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hildén KS, Hope R, Hossain A, Karabika E, Karaffa L, Karányi Z, Kraševec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, et al. . 2017. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18:28. doi:10.1186/s13059-017-1151-0. - DOI - PMC - PubMed
-
- Alberts AW, Chen J, Kuron G, Hunt V, Huff J, Hoffman C, Rothrock J, Lopez M, Joshua H, Harris E, Patchett A, Monaghan R, Currie S, Stapley E, Albers-Schonberg G, Hensens O, Hirshfield J, Hoogsteen K, Liesch J, Springer J. 1980. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci U S A 77:3957–3961. doi:10.1073/pnas.77.7.3957. - DOI - PMC - PubMed
-
- Pontecorvo G, Roper JA, Chemmons LM, Macdonald KD, Bufton AWJ. 1953. The genetics of Aspergillus nidulans, p 141–238. In Demerec M. (ed), Advances in genetics. Academic Press, Cambridge, MA. - PubMed
-
- Challa S. 2018. Pathogenesis and pathology of invasive aspergillosis. Curr Fungal Infect Rep 12:23–32. doi:10.1007/s12281-018-0310-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

