16S rRNA deep sequencing for the characterization of healthy human pharyngeal microbiome
- PMID: 31213755
- PMCID: PMC6528694
16S rRNA deep sequencing for the characterization of healthy human pharyngeal microbiome
Abstract
Background: The recent advent of high-throughput sequencing methods enabled the study of the composition of the upper respiratory tract (URT) microbial ecosystem and its relationship with health and disease in immense detail. The aim of the present study was the characterization of the human pharyngeal microbiome of healthy individuals in Greece.
Materials and methods: We obtained ten pharyngeal specimens from healthy volunteers, Greek resident, with Greek nationality, who were eligible to the selection criteria. The construction of DNA libraries was performed by using two primer sets that amplify selectively the corresponding hypervariable regions of the 16s region in bacteria (V2-V9). The Ion Torrent PGM platform was used for the performance of next-generation sequencing.
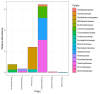
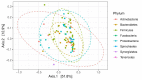
Results: In the study samples, twelve phyla were identified. The most abundant ones were Firmicutes, Proteobacteria, Bacteroidetes, followed by Actinobacteria and Fusobacteria. Seventy-nine families, 79 genera and 137 species were identified and characterized. Moreover, 17 unique differentially abundant families, 30 unique differentially abundant genera and 24 unique differentially abundant species were identified among healthy subgroups with adjusted p-values <0.05. At the genus level, Moraxella (Proteobacteria) and Gemella (Firmicutes) were detected with a statistical significance in non-smokers, while Bifidobacterium (Actinobacteria), Alloscardovia (Actinobacteria), Dialister (Firmicutes) and Filifactor (Firmicutes) were present mostly in smokers.
Conclusions: The URT is colonized by a variety of protective and potentially pathogenic bacteria. This microbiome system is highly diverse and varies significantly between individuals. Geographic location and ethnicity are considered to be a strong determinants and factors affecting the diversity and abundance of the URT microbiome. Although some of the most abundant families are common irrespective of these factors, the dominance patterns are usually different between the study subjects and between the studies from other geographic locations. Unique differentially abundant families, genera and species were identified, and further studies are needed to elucidate their role. Further studies should focus on the investigation of the URT microbiome dynamics and the interaction with the host in health and disease. HIPPOKRATIA 2018, 22(1): 29-36.
Keywords: 16srRNA; Greece; Pharyngeal microbiome; healthy; sequencing.
Conflict of interest statement
Authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
